8R61
 
 | |
8ONN
 
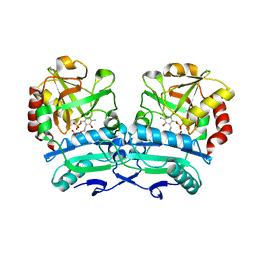 | | Crystal structure of D-amino acid aminotransferase from Aminobacterium colombiense point mutant E113A complexed with 3-aminooxypropionic acid | | Descriptor: | 3-[(~{E})-[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]methylideneamino]oxypropanoic acid, Aminotransferase class IV | | Authors: | Matyuta, I.O, Boyko, K.M, Minyaev, M.E, Shilova, S.A, Bezsudnova, E.Y, Popov, V.O. | | Deposit date: | 2023-04-03 | | Release date: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | In search for structural targets for engineering d-amino acid transaminase: modulation of pH optimum and substrate specificity.
Biochem.J., 480, 2023
|
|
7QG9
 
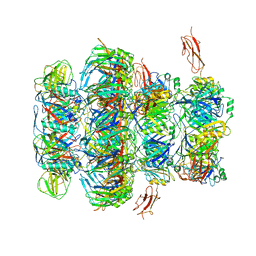 | | Tail tip of siphophage T5 : common core proteins | | Descriptor: | Distal tail protein, L-shaped tail fiber protein p132, Minor tail protein, ... | | Authors: | Linares, R, Arnaud, C.A, Effantin, G, Darnault, C, Epalle, N, Boeri Erba, E, Schoehn, G, Breyton, C. | | Deposit date: | 2021-12-07 | | Release date: | 2022-12-21 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Structural basis of bacteriophage T5 infection trigger and E. coli cell wall perforation.
Sci Adv, 9, 2023
|
|
6MYQ
 
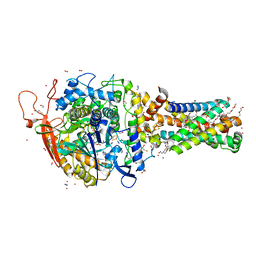 | | Avian mitochondrial complex II with ferulenol bound | | Descriptor: | (~{Z})-2-oxidanylbut-2-enedioic acid, 1,2-Distearoyl-sn-glycerophosphoethanolamine, 4-oxidanyl-3-[(2~{E},6~{E})-3,7,11-trimethyldodeca-2,6,10-trienyl]chromen-2-one, ... | | Authors: | Berry, E.A, Huang, L.-S. | | Deposit date: | 2018-11-02 | | Release date: | 2019-11-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Crystallographic investigation of the ubiquinone binding site of respiratory Complex II and its inhibitors.
Biochim Biophys Acta Proteins Proteom, 1869, 2021
|
|
8TL4
 
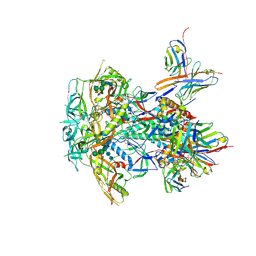 | | CRYO-EM STRUCTURE OF HIV-1 BG505DS-SOSIP.664 ENV TRIMER BOUND TO DJ85-e.01 FAB | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BG505 DS-SOSIP Surface protein gp120, ... | | Authors: | Pletnev, S, Hoyt, F, Fischer, E, Kwong, P. | | Deposit date: | 2023-07-26 | | Release date: | 2024-08-28 | | Last modified: | 2025-01-15 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Potent and broad HIV-1 neutralization in fusion peptide-primed SHIV-infected macaques.
Cell, 187, 2024
|
|
6GS1
 
 | | Crystal structure of peptide transporter DtpA-nanobody in MES buffer | | Descriptor: | Dipeptide and tripeptide permease A, Nanobody 00 | | Authors: | Ural-Blimke, Y, Flayhan, A, Loew, C, Quistgaard, E.M. | | Deposit date: | 2018-06-13 | | Release date: | 2019-01-30 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.29 Å) | | Cite: | Structure of Prototypic Peptide Transporter DtpA from E. coli in Complex with Valganciclovir Provides Insights into Drug Binding of Human PepT1.
J. Am. Chem. Soc., 141, 2019
|
|
8VXL
 
 | | Crystal Structure of the External Aldimine Complex of Pyridoxal-5'-Tetrazole and (S)-4-amino-5-phenoxypentanoate with the Bacillus subtilis GabR C-terminal Effector-binding and Oligomerization Domain | | Descriptor: | (4S)-4-[(E)-({3-hydroxy-2-methyl-5-[(E)-2-(1H-tetrazol-5-yl)ethenyl]pyridin-4-yl}methylidene)amino]-5-phenoxypentanoic acid, GLYCEROL, HTH-type transcriptional regulatory protein GabR, ... | | Authors: | Kaley, N.E, Liveris, Z.J, Becker, D.P, Liu, D. | | Deposit date: | 2024-02-05 | | Release date: | 2025-01-08 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Bioisosteric replacement of pyridoxal-5'-phosphate to pyridoxal-5'-tetrazole targeting Bacillus subtilis GabR.
Protein Sci., 34, 2025
|
|
6GS7
 
 | | Crystal structure of peptide transporter DtpA-nanobody in glycine buffer | | Descriptor: | Dipeptide and tripeptide permease A, nanobody | | Authors: | Ural-Blimke, Y, Flayhan, A, Quistgaard, E.M, Loew, C. | | Deposit date: | 2018-06-13 | | Release date: | 2019-01-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure of Prototypic Peptide Transporter DtpA from E. coli in Complex with Valganciclovir Provides Insights into Drug Binding of Human PepT1.
J. Am. Chem. Soc., 141, 2019
|
|
6GZ5
 
 | | tRNA translocation by the eukaryotic 80S ribosome and the impact of GTP hydrolysis, Translocation-intermediate-POST-3 (TI-POST-3) | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 5.8S ribosomal RNA, ... | | Authors: | Flis, J, Holm, M, Rundlet, E.J, Loerke, J, Hilal, T, Dabrowski, M, Buerger, J, Mielke, T, Blanchard, S.C, Spahn, C.M.T, Budkevich, T.V. | | Deposit date: | 2018-07-03 | | Release date: | 2018-12-05 | | Last modified: | 2022-03-30 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | tRNA Translocation by the Eukaryotic 80S Ribosome and the Impact of GTP Hydrolysis.
Cell Rep, 25, 2018
|
|
8EF6
 
 | | Morphine-bound mu-opioid receptor-Gi complex | | Descriptor: | (7R,7AS,12BS)-3-METHYL-2,3,4,4A,7,7A-HEXAHYDRO-1H-4,12-METHANO[1]BENZOFURO[3,2-E]ISOQUINOLINE-7,9-DIOL, CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Zhuang, Y, Wang, Y, Guo, S, Zhou, X.E, Rao, Q, He, X, He, B, Liu, J, Zhou, Q, Wang, X, Liu, W, Jiang, X, Yang, D, Chen, X, Jiang, Y, Jiang, H, Shen, J, Melcher, K, Wang, M, Xie, X, Xu, H.E. | | Deposit date: | 2022-09-08 | | Release date: | 2022-11-09 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Molecular recognition of morphine and fentanyl by the human mu-opioid receptor.
Cell, 185, 2022
|
|
3FFZ
 
 | | Domain organization in Clostridium butulinum neurotoxin type E is unique: Its implication in faster translocation | | Descriptor: | ACETATE ION, Botulinum neurotoxin type E, SODIUM ION, ... | | Authors: | Kumaran, D, Eswaramoorthy, S, Swaminathan, S. | | Deposit date: | 2008-12-04 | | Release date: | 2008-12-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Domain organization in Clostridium botulinum neurotoxin type E is unique: its implication in faster translocation.
J.Mol.Biol., 386, 2009
|
|
8FE8
 
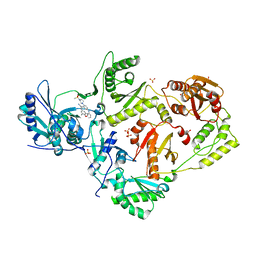 | | Crystal Structure of HIV-1 RT in Complex with the non-nucleoside inhibitor 18b1 | | Descriptor: | 1,2-ETHANEDIOL, 5-[(4-{4-[(E)-2-cyanoethenyl]-2,6-dimethylphenoxy}pyrimidin-2-yl)amino]-2-[4-(methanesulfonyl)piperazin-1-yl]benzonitrile, Reverse transcriptase p51, ... | | Authors: | Rumrill, S, Ruiz, F.X, Arnold, E. | | Deposit date: | 2022-12-05 | | Release date: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery of diarylpyrimidine derivatives bearing piperazine sulfonyl as potent HIV-1 nonnucleoside reverse transcriptase inhibitors.
Commun Chem, 6, 2023
|
|
6EWK
 
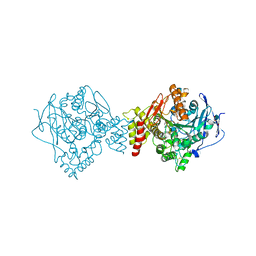 | | T. californica AChE in complex with a 3-hydroxy-2-pyridine aldoxime. | | Descriptor: | 2-[(~{E})-hydroxyiminomethyl]-6-(5-morpholin-4-ylpentyl)pyridin-3-ol, 2-acetamido-2-deoxy-beta-D-glucopyranose, Acetylcholinesterase, ... | | Authors: | de la Mora, E, Weik, M, Braiki, A, Mougeot, R, Jean, L, Renard, P.I. | | Deposit date: | 2017-11-04 | | Release date: | 2018-11-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Potent 3-Hydroxy-2-Pyridine Aldoxime Reactivators of Organophosphate-Inhibited Cholinesterases with Predicted Blood-Brain Barrier Penetration.
Chemistry, 24, 2018
|
|
8J6N
 
 | | Crystal structure of Cystathionine gamma-lyase in complex with compound 1 | | Descriptor: | 1,2-ETHANEDIOL, Cystathionine gamma-lyase, GLYCEROL, ... | | Authors: | Hibi, R, Toma-Fukai, S, Shimizu, T, Hanaoka, K. | | Deposit date: | 2023-04-26 | | Release date: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of a cystathionine gamma-lyase (CSE) selective inhibitor targeting active-site pyridoxal 5'-phosphate (PLP) via Schiff base formation.
Sci Rep, 13, 2023
|
|
7PH4
 
 | | AMP-PNP bound nanodisc reconstituted MsbA with nanobodies, spin-labeled at position T68C | | Descriptor: | (1~{R},4~{R},11~{S},14~{S},19~{Z})-19-[2-[2,5-bis(oxidanylidene)pyrrolidin-1-yl]ethylimino]-7,8,17,18-tetraoxa-1,4,11,14-tetrazatricyclo[12.6.2.2^{4,11}]tetracosane-6,9,16-trione, ATP-dependent lipid A-core flippase, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Parey, K, Januliene, D, Galazzo, L, Meier, G, Vecchis, D, Striednig, B, Hilbi, H, Schaefer, L.V, Kuprov, I, Bordignon, E, Seeger, M.A, Moeller, A. | | Deposit date: | 2021-08-16 | | Release date: | 2022-08-24 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | The ABC transporter MsbA adopts the wide inward-open conformation in E. coli cells.
Sci Adv, 8, 2022
|
|
8PMX
 
 | |
7SN4
 
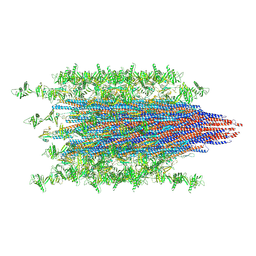 | |
7SN7
 
 | |
3E1Q
 
 | | Crystal structure of W133F variant E. coli Bacterioferritn with iron. | | Descriptor: | BACTERIOFERRITIN, FE (II) ION, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Crow, A, Lawson, T.L, Lewin, A, Moore, G.R, Le Brun, N.E. | | Deposit date: | 2008-08-04 | | Release date: | 2009-08-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Monitoring the iron status of the ferroxidase center of Escherichia coli bacterioferritin using fluorescence spectroscopy.
Biochemistry, 48, 2009
|
|
8DO1
 
 | | Cryo-EM structure of the human Sec61 complex inhibited by ipomoeassin F | | Descriptor: | Protein transport protein Sec61 subunit alpha isoform 1, Protein transport protein Sec61 subunit beta, Protein transport protein Sec61 subunit gamma, ... | | Authors: | Park, E, Itskanov, S. | | Deposit date: | 2022-07-12 | | Release date: | 2023-05-24 | | Last modified: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | A common mechanism of Sec61 translocon inhibition by small molecules.
Nat.Chem.Biol., 19, 2023
|
|
6G4M
 
 | | Torpedo californica acetylcholinesterase bound to uncharged hybrid reactivator 1 | | Descriptor: | 2-[(~{E})-hydroxyiminomethyl]-6-[4-(1,2,3,4-tetrahydroacridin-9-ylamino)butyl]pyridin-3-ol, 2-acetamido-2-deoxy-beta-D-glucopyranose, Acetylcholinesterase, ... | | Authors: | Santoni, G, De la Mora, E, de Souza, J, Silman, I, Sussman, J, Baati, R, Weik, M, Nachon, F. | | Deposit date: | 2018-03-28 | | Release date: | 2018-08-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Structure-Based Optimization of Nonquaternary Reactivators of Acetylcholinesterase Inhibited by Organophosphorus Nerve Agents.
J. Med. Chem., 61, 2018
|
|
8GS2
 
 | | Structure of the Cas7-11-Csx29-guide RNA-target RNA (non-matching PFS) complex | | Descriptor: | ADENOSINE MONOPHOSPHATE, CHAT domain-containing protein, CRISPR-associated RAMP family protein, ... | | Authors: | Kato, K, Okazaki, S, Ishikawa, J, Isayama, Y, Nishizawa, T, Nishimasu, H. | | Deposit date: | 2022-09-04 | | Release date: | 2022-11-09 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | RNA-triggered protein cleavage and cell growth arrest by the type III-E CRISPR nuclease-protease.
Science, 378, 2022
|
|
6G4O
 
 | | Non-aged form of Torpedo californica acetylcholinesterase inhibited by tabun analog NEDPA bound to uncharged reactivator 1 | | Descriptor: | 2-[(~{E})-hydroxyiminomethyl]-6-[4-(1,2,3,4-tetrahydroacridin-9-ylamino)butyl]pyridin-3-ol, 2-acetamido-2-deoxy-beta-D-glucopyranose, Acetylcholinesterase, ... | | Authors: | Santoni, G, De la Mora, E, de Souza, J, Silman, I, Sussman, J, Baati, R, Weik, M, Nachon, F. | | Deposit date: | 2018-03-28 | | Release date: | 2018-08-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Structure-Based Optimization of Nonquaternary Reactivators of Acetylcholinesterase Inhibited by Organophosphorus Nerve Agents.
J. Med. Chem., 61, 2018
|
|
8CBA
 
 | |
6OVB
 
 | | Crystal structure of a Bacillus thuringiensis Cry1Da tryptic core variant | | Descriptor: | Active core crystal toxin protein 1D | | Authors: | Rydel, T.J, Halls, C, Evdokimov, A.G, Beishir, S.C. | | Deposit date: | 2019-05-07 | | Release date: | 2019-06-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.611 Å) | | Cite: | Bacillus thuringiensis Cry1Da_7 and Cry1B.868 Protein Interactions with Novel Receptors Allow Control of Resistant Fall Armyworms, Spodoptera frugiperda (J.E. Smith).
Appl.Environ.Microbiol., 85, 2019
|
|
