3MSL
 
 | |
3MSN
 
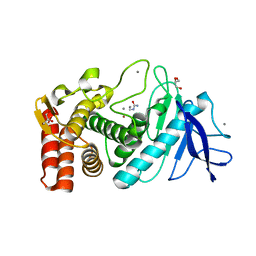 | | Crystal structure of Thermolysin in complex with N-methylurea | | Descriptor: | CALCIUM ION, GLYCEROL, N-METHYLUREA, ... | | Authors: | Behnen, J, Heine, A, Klebe, G. | | Deposit date: | 2010-04-29 | | Release date: | 2011-05-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Experimental and computational active site mapping as a starting point to fragment-based lead discovery.
Chemmedchem, 7, 2012
|
|
3MSO
 
 | |
3MSP
 
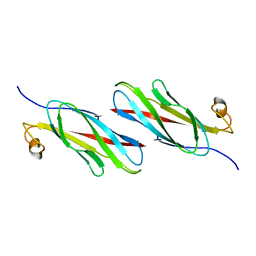 | | MOTILE MAJOR SPERM PROTEIN (MSP) OF ASCARIS SUUM, NMR, 20 STRUCTURES | | Descriptor: | MAJOR SPERM PROTEIN | | Authors: | Haaf, A, Leclaire III, L, Roberts, G, Kent, H.M, Roberts, T.M, Stewart, M, Neuhaus, D. | | Deposit date: | 1998-09-10 | | Release date: | 1999-04-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the motile major sperm protein (MSP) of Ascaris suum - evidence for two manganese binding sites and the possible role of divalent cations in filament formation.
J.Mol.Biol., 284, 1998
|
|
3MSQ
 
 | |
3MSR
 
 | |
3MSS
 
 | |
3MST
 
 | |
3MSU
 
 | | Crystal Structure of Citrate Synthase from Francisella tularensis | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETIC ACID, CHLORIDE ION, ... | | Authors: | Maltseva, N, Kim, Y, Hasseman, J, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-04-29 | | Release date: | 2010-05-26 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.843 Å) | | Cite: | Crystal Structure of Citrate Synthase from Francisella tularensis
To be Published
|
|
3MSV
 
 | |
3MSW
 
 | |
3MSX
 
 | | Crystal structure of RhoA.GDP.MgF3 in complex with GAP domain of ArhGAP20 | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Rho GTPase-activating protein 20, ... | | Authors: | Utepbergenov, D, Cooper, D.R, Derewenda, U, Derewenda, Z.S. | | Deposit date: | 2010-04-29 | | Release date: | 2011-03-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Mechanism of molecular specificity of RhoGAP domains towards small GTPases of RhoA family.
To be Published
|
|
3MSY
 
 | |
3MSZ
 
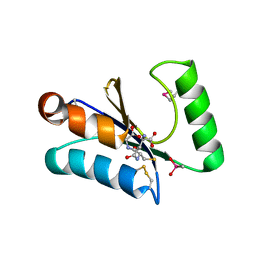 | | Crystal Structure of Glutaredoxin 1 from Francisella tularensis Complexed with Cacodylate | | Descriptor: | CACODYLATE ION, GLUTATHIONE, GLYCEROL, ... | | Authors: | Maltseva, N, Kim, Y, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-04-29 | | Release date: | 2010-05-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.053 Å) | | Cite: | Crystal Structure of Glutaredoxin 1 from Francisella tularensis Complexed with Cacodylate
To be Published
|
|
3MT0
 
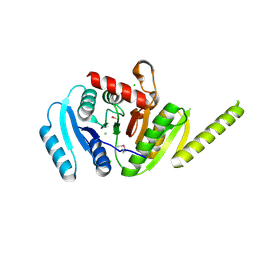 | | The crystal structure of a functionally unknown protein PA1789 from Pseudomonas aeruginosa PAO1 | | Descriptor: | CHLORIDE ION, uncharacterized protein PA1789 | | Authors: | Tan, K, Chang, C, Tesar, C, Bearden, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-04-29 | | Release date: | 2010-05-12 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.582 Å) | | Cite: | The crystal structure of a functionally unknown protein PA1789 from Pseudomonas aeruginosa PAO1
To be Published
|
|
3MT1
 
 | | Crystal structure of putative carboxynorspermidine decarboxylase protein from Sinorhizobium meliloti | | Descriptor: | Putative carboxynorspermidine decarboxylase protein, SULFATE ION | | Authors: | Chang, C, Xu, X, Cui, H, Chin, S, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-04-29 | | Release date: | 2010-06-30 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of putative carboxynorspermidine decarboxylase protein from Sinorhizobium meliloti
To be Published
|
|
3MT5
 
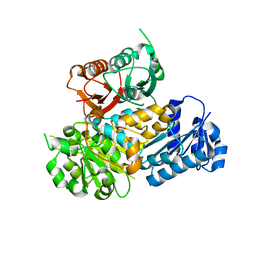 | | Crystal Structure of the Human BK Gating Apparatus | | Descriptor: | CALCIUM ION, Potassium large conductance calcium-activated channel, subfamily M, ... | | Authors: | Yuan, P, MacKinnon, R. | | Deposit date: | 2010-04-30 | | Release date: | 2010-06-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of the human BK channel Ca2+-activation apparatus at 3.0 A resolution.
Science, 329, 2010
|
|
3MT6
 
 | | Structure of ClpP from Escherichia coli in complex with ADEP1 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACYLDEPSIPEPTIDE 1, ATP-dependent Clp protease proteolytic subunit | | Authors: | Chung, Y.S. | | Deposit date: | 2010-04-30 | | Release date: | 2010-11-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Acyldepsipeptide antibiotics induce the formation of a structured axial channel in ClpP: A model for the ClpX/ClpA-bound state of ClpP.
Chem.Biol., 17, 2010
|
|
3MT7
 
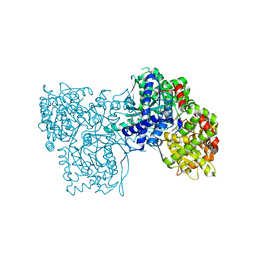 | |
3MT8
 
 | |
3MT9
 
 | |
3MTA
 
 | |
3MTB
 
 | |
3MTC
 
 | | Crystal Structure of INPP5B in complex with phosphatidylinositol 4-phosphate | | Descriptor: | (2R)-3-{[(S)-hydroxy{[(1R,2R,3R,4R,5S,6R)-2,3,5,6-tetrahydroxy-4-(phosphonooxy)cyclohexyl]oxy}phosphoryl]oxy}propane-1,2-diyl dioctanoate, CHLORIDE ION, GLYCEROL, ... | | Authors: | Tresaugues, L, Welin, M, Arrowsmith, C.H, Berglund, H, Bountra, C, Collins, R, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, I, Karlberg, T, Kol, S, Kotenyova, T, Moche, M, Nyman, T, Persson, C, Schuler, H, Schutz, P, Siponen, M.I, Thorsell, A.G, van der Berg, S, Wahlberg, E, Weigelt, J, Wisniewska, M, Nordlund, P, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-04-30 | | Release date: | 2010-06-30 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for phosphoinositide substrate recognition, catalysis, and membrane interactions in human inositol polyphosphate 5-phosphatases
Structure, 22, 2014
|
|
3MTD
 
 | |
