4YJF
 
 | | Crystal structure of DAAO(Y228L/R283G) variant (S-methylbenzylamine binding form) | | Descriptor: | (1S)-1-phenylethanamine, D-amino-acid oxidase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Nakano, S, Yasukawa, K, Kawahara, N, Ishitsubo, E, Tokiwa, H, Asano, Y. | | Deposit date: | 2015-03-03 | | Release date: | 2016-04-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of DAAO variant
To Be Published
|
|
9CXR
 
 | | X-ray Crystallographic Structure of the Poly(Hexamethylene Adipamide) (Nylon66) Hydrolase Nyl50 at Room Temperature | | Descriptor: | 1,2-ETHANEDIOL, Poly(hexamethylene adipamide) hydrolase, SODIUM ION, ... | | Authors: | Meilleur, F, Williams, A.N, Drufva, E.E, Parks, J.M, Chen, S.H, Michener, J.K. | | Deposit date: | 2024-07-31 | | Release date: | 2025-01-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Identification and characterization of substrate- and product-selective nylon hydrolases.
Biorxiv, 2024
|
|
4YJG
 
 | | Crystal structure of DAAO(Y228L/R283G) variant (R-3-amino 1-phenylbutane binding form) | | Descriptor: | (2R)-4-phenylbutan-2-amine, D-amino-acid oxidase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Nakano, S, Yasukawa, K, Kawahara, N, Ishitsubo, E, Tokiwa, H, Asano, Y. | | Deposit date: | 2015-03-03 | | Release date: | 2016-04-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of DAAO variant
To Be Published
|
|
4WUN
 
 | | Structure of FGFR1 in complex with AZD4547 (N-{3-[2-(3,5-DIMETHOXYPHENYL)ETHYL]-1H-PYRAZOL-5-YL}-4-[(3R,5S)-3,5-DIMETHYLPIPERAZIN-1-YL]BENZAMIDE) at 1.65 angstrom | | Descriptor: | Fibroblast growth factor receptor 1, N-{3-[2-(3,5-dimethoxyphenyl)ethyl]-1H-pyrazol-5-yl}-4-[(3R,5S)-3,5-dimethylpiperazin-1-yl]benzamide | | Authors: | Squire, C.J, Yosaatmadja, C.J. | | Deposit date: | 2014-11-02 | | Release date: | 2014-11-19 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The 1.65 angstrom resolution structure of the complex of AZD4547 with the kinase domain of FGFR1 displays exquisite molecular recognition.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4X4A
 
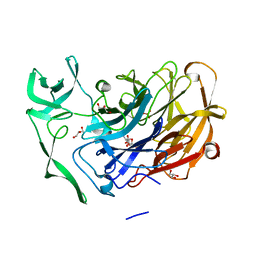 | | Crystal structure of the intramolecular trans-sialidase from Ruminococcus gnavus in complex with 2,7-Anhydro-Neu5Ac | | Descriptor: | 2-ACETYLAMINO-7-(1,2-DIHYDROXY-ETHYL)-3-HYDROXY-6,8-DIOXA-BICYCLO[3.2.1]OCTANE-5-CARBOXYLIC ACID, ACETYL GROUP, Anhydrosialidase, ... | | Authors: | Owen, C.D, Tailford, L.E, Taylor, G.L, Juge, N. | | Deposit date: | 2014-12-02 | | Release date: | 2015-07-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Discovery of intramolecular trans-sialidases in human gut microbiota suggests novel mechanisms of mucosal adaptation.
Nat Commun, 6, 2015
|
|
4WNA
 
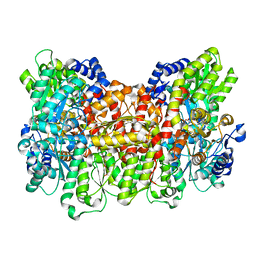 | | Structure of the Nitrogenase MoFe Protein from Azotobacter vinelandii Pressurized with Xenon | | Descriptor: | 3-HYDROXY-3-CARBOXY-ADIPIC ACID, FE (III) ION, FE(8)-S(7) CLUSTER, ... | | Authors: | Morrison, C.N, Hoy, J.A, Zhang, L, Einsle, O, Rees, D.C. | | Deposit date: | 2014-10-11 | | Release date: | 2015-03-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Substrate Pathways in the Nitrogenase MoFe Protein by Experimental Identification of Small Molecule Binding Sites.
Biochemistry, 54, 2015
|
|
4WPH
 
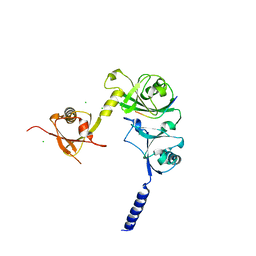 | |
4WR3
 
 | | Y274F alanine racemase from E. coli | | Descriptor: | Alanine racemase, biosynthetic, GLYCEROL, ... | | Authors: | Squire, C.J, Yosaatmadja, Y, Patrick, W.M. | | Deposit date: | 2014-10-23 | | Release date: | 2015-11-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mechanistic and Evolutionary Insights from the Reciprocal Promiscuity of Two Pyridoxal Phosphate-dependent Enzymes.
J.Biol.Chem., 291, 2016
|
|
9DP0
 
 | | Fibrillar assembly of racemic, C-alpha methylated, macrocyclic beta-hairpins | | Descriptor: | CITRATE ANION, PENTAETHYLENE GLYCOL, racemic, ... | | Authors: | Samdin, T.D, Lubkowski, J, Anderson, C.F, Schneider, J.P. | | Deposit date: | 2024-09-20 | | Release date: | 2025-04-30 | | Last modified: | 2025-05-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | From Hydrogel to Crystal: A Molecular Design Strategy that Chemically Modifies Racemic Gel-Forming Peptides to Furnish Crystalline Fibrils Stabilized by Parallel Rippled beta-Sheets.
J.Am.Chem.Soc., 147, 2025
|
|
4YEI
 
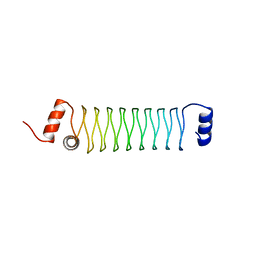 | | Beta1mut synthetic solenoid protein | | Descriptor: | beta1mut | | Authors: | Murray, J.W, Kraatz, S. | | Deposit date: | 2015-02-24 | | Release date: | 2016-03-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.55 Å) | | Cite: | Synthetic beta-solenoid proteins with the fragment-free computational design of a beta-hairpin extension.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
9DOX
 
 | | Fibrillar assembly of an L-enantiopure, C-alpha methylated, macrocyclic beta-hairpin | | Descriptor: | 5-amino-2,4,6-triiodobenzene-1,3-dicarboxylic acid, L-enantiopure, C-alpha methylated, ... | | Authors: | Samdin, T.D, Lubkowski, J, Anderson, C.F, Schneider, J.P. | | Deposit date: | 2024-09-20 | | Release date: | 2025-04-30 | | Last modified: | 2025-05-07 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | From Hydrogel to Crystal: A Molecular Design Strategy that Chemically Modifies Racemic Gel-Forming Peptides to Furnish Crystalline Fibrils Stabilized by Parallel Rippled beta-Sheets.
J.Am.Chem.Soc., 147, 2025
|
|
9DOY
 
 | | Fibrillar assembly of an D-enantiopure, C-alpha methylated, macrocyclic beta-hairpin | | Descriptor: | 5-amino-2,4,6-triiodobenzene-1,3-dicarboxylic acid, D-enantiopure, C-alpha methylated, ... | | Authors: | Samdin, T.D, Lubkowski, J, Anderson, C.F, Schneider, J.P. | | Deposit date: | 2024-09-20 | | Release date: | 2025-04-30 | | Last modified: | 2025-05-07 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | From Hydrogel to Crystal: A Molecular Design Strategy that Chemically Modifies Racemic Gel-Forming Peptides to Furnish Crystalline Fibrils Stabilized by Parallel Rippled beta-Sheets.
J.Am.Chem.Soc., 147, 2025
|
|
8W6W
 
 | | Crystal Structure of C-terminal domain of nucleocapsid protein from SARS-CoV-2 in complex with ampicillin | | Descriptor: | (2S,5R,6R)-6-{[(2R)-2-AMINO-2-PHENYLETHANOYL]AMINO}-3,3-DIMETHYL-7-OXO-4-THIA-1-AZABICYCLO[3.2.0]HEPTANE-2-CARBOXYLIC ACID, 1,2-ETHANEDIOL, Nucleoprotein | | Authors: | Dhaka, P, Mahto, J.K, Tomar, S, Kumar, P. | | Deposit date: | 2023-08-30 | | Release date: | 2025-03-05 | | Last modified: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural insights into the RNA binding inhibitors of the C-terminal domain of the SARS-CoV-2 nucleocapsid.
J.Struct.Biol., 217, 2025
|
|
4YW1
 
 | | Crystal Structure of Streptococcus pneumoniae NanC, complex with Neu5Ac and Neu5Ac2en following soaking with 3'SL | | Descriptor: | 2-DEOXY-2,3-DEHYDRO-N-ACETYL-NEURAMINIC ACID, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Owen, C.D, Lukacik, P, Potter, J.A, Walsh, M, Taylor, G.L. | | Deposit date: | 2015-03-20 | | Release date: | 2015-09-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Streptococcus pneumoniae NanC: STRUCTURAL INSIGHTS INTO THE SPECIFICITY AND MECHANISM OF A SIALIDASE THAT PRODUCES A SIALIDASE INHIBITOR.
J.Biol.Chem., 290, 2015
|
|
4YW3
 
 | | Crystal Structure of Streptococcus pneumoniae NanC, complex with Neu5Ac and Neu5Ac2en following soaking with Neu5Ac2en | | Descriptor: | 2-DEOXY-2,3-DEHYDRO-N-ACETYL-NEURAMINIC ACID, GLYCEROL, N-acetyl-alpha-neuraminic acid, ... | | Authors: | Owen, C.D, Lukacik, P, Potter, J.A, Walsh, M, Taylor, G.L. | | Deposit date: | 2015-03-20 | | Release date: | 2015-09-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Streptococcus pneumoniae NanC: STRUCTURAL INSIGHTS INTO THE SPECIFICITY AND MECHANISM OF A SIALIDASE THAT PRODUCES A SIALIDASE INHIBITOR.
J.Biol.Chem., 290, 2015
|
|
9BLJ
 
 | |
4X6K
 
 | | Crystal structure of the intramolecular trans-sialidase from Ruminococcus gnavus in complex with Siastatin B | | Descriptor: | (2S,3R,4S,5S)-2-(acetylamino)-5-carboxy-3,4-dihydroxypiperidinium, ACETYL GROUP, Anhydrosialidase | | Authors: | Owen, C.D, Tailford, L.E, Taylor, G.L, Juge, N. | | Deposit date: | 2014-12-08 | | Release date: | 2015-07-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Discovery of intramolecular trans-sialidases in human gut microbiota suggests novel mechanisms of mucosal adaptation.
Nat Commun, 6, 2015
|
|
4WRB
 
 | | Macrophage Migration Inhibitory Factor in complex with a biaryltriazole inhibitor (3b-190) | | Descriptor: | 4-{4-[6-(2-methoxyethoxy)quinolin-2-yl]-1H-1,2,3-triazol-1-yl}phenol, GLYCEROL, ISOPROPYL ALCOHOL, ... | | Authors: | Robertson, M.J, Baxter, R.H.G, Jorgensen, W.L. | | Deposit date: | 2014-10-23 | | Release date: | 2015-03-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Design, synthesis, and protein crystallography of biaryltriazoles as potent tautomerase inhibitors of macrophage migration inhibitory factor.
J.Am.Chem.Soc., 137, 2015
|
|
4WX2
 
 | | Crystal structure of Tryptophan Synthase from Salmonella typhimurium in complex with two F6F molecules in the alpha-site and one F6F molecule in the beta-site | | Descriptor: | 1,2-ETHANEDIOL, 2-{[4-(TRIFLUOROMETHOXY)BENZOYL]AMINO}ETHYL DIHYDROGEN PHOSPHATE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Hilario, E, Caulkins, B.G, Young, R.P, Niks, D, Dunn, M.F, Mueller, L.J, Fan, L. | | Deposit date: | 2014-11-13 | | Release date: | 2015-11-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Visualizing the tunnel in tryptophan synthase with crystallography: Insights into a selective filter for accommodating indole and rejecting water.
Biochim.Biophys.Acta, 1864, 2016
|
|
4XZK
 
 | |
4YPG
 
 | |
9CM8
 
 | | UDP-GlcNAc 2-epimerase MnaA of Paenibacillus alvei | | Descriptor: | DI(HYDROXYETHYL)ETHER, UDP-N-acetylglucosamine 2-epimerase (non-hydrolyzing) | | Authors: | Legg, M.S.G, Mateyko, N, Evans, S.V. | | Deposit date: | 2024-07-13 | | Release date: | 2024-10-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Insights into structure and activity of a UDP-GlcNAc 2-epimerase involved in secondary cell wall polymer biosynthesis in Paenibacillus alvei.
Front Mol Biosci, 11, 2024
|
|
4YIP
 
 | | X-ray structure of the iron/manganese cambialistic superoxide dismutase from Streptococcus mutans | | Descriptor: | FE (III) ION, Superoxide dismutase [Mn/Fe] | | Authors: | Russo Krauss, I, Merlino, A, Pica, A, Sica, F. | | Deposit date: | 2015-03-02 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Fine tuning of metal-specific activity in the Mn-like group of cambialistic superoxide dismutases
Rsc Adv, 5, 2015
|
|
4YNQ
 
 | | TREX1-dsDNA complex | | Descriptor: | DNA (24-MER), DNA (5'-D(P*GP*TP*GP*CP*TP*GP*AP*CP*GP*TP*CP*AP*GP*CP*AP*CP*GP*AP*CP*G)-3'), MAGNESIUM ION, ... | | Authors: | Fye, J.M, Harvey, S, Perrino, F.W, Hollis, T. | | Deposit date: | 2015-03-10 | | Release date: | 2015-05-27 | | Last modified: | 2025-02-12 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Exonuclease TREX1 degrades double-stranded DNA to prevent spontaneous lupus-like inflammatory disease.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4Z41
 
 | | X-ray structure of the adduct formed in the reaction between lysozyme and a platinum(II) Compound with a S,O Bidentate Ligand (9a=Chloro-(1-(3'-hydroxy)-3-(methylthio)-3-thioxo-prop-1-en-1-olate-O,S)-(dimethylsulfoxide-S)-platinum(II)) | | Descriptor: | 1,2-ETHANEDIOL, 3-[2-chloranyl-2-[dimethyl(oxidanyl)-{4}-sulfanyl]-4-ethylsulfanyl-1-oxa-3{3}-thia-2{4}-platinacyclohexa-3,5-dien-6-yl]phenol, DIMETHYL SULFOXIDE, ... | | Authors: | Merlino, A. | | Deposit date: | 2015-04-01 | | Release date: | 2015-09-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Platinum(II) Complexes with O,S Bidentate Ligands: Biophysical Characterization, Antiproliferative Activity, and Crystallographic Evidence of Protein Binding.
Inorg.Chem., 54, 2015
|
|
