9PQ7
 
 | | MBP-Mcl1 in complex with ligand 21b | | Descriptor: | 1,2-ETHANEDIOL, 17-chloranyl-33-fluoranyl-12-[2-(2-methoxyethoxy)ethyl]-5,14,22-trimethyl-28-oxa-9-thia-5,6,12,13,24-pentazaheptacyclo[27.7.1.1^{4,7}.0^{11,15}.0^{16,21}.0^{20,24}.0^{30,35}]octatriaconta-1(36),4(38),6,11(15),13,16,18,20,22,29(37),30(35),31,33-tridecaene-23-carboxylic acid, FORMIC ACID, ... | | Authors: | Miller, B.R, Shaffer, P. | | Deposit date: | 2025-07-22 | | Release date: | 2025-10-08 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | In Pursuit of Best-in-Class MCL-1 Inhibitors: Discovery of Highly Potent 1,4-Indoyl Macrocycles with Favorable Physicochemical Properties.
J.Med.Chem., 68, 2025
|
|
2HMX
 
 | | HUMAN IMMUNODEFICIENCY VIRUS TYPE 1 MATRIX PROTEIN | | Descriptor: | HUMAN IMMUNODEFICIENCY VIRUS TYPE 1 MATRIX PROTEIN | | Authors: | Massiah, M.A, Starich, M.R, Paschall, C, Christensen, A.M, Sundquist, W.I, Summers, M.F. | | Deposit date: | 1995-09-22 | | Release date: | 1996-01-29 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure of the human immunodeficiency virus type 1 matrix protein.
J.Mol.Biol., 244, 1994
|
|
3RG1
 
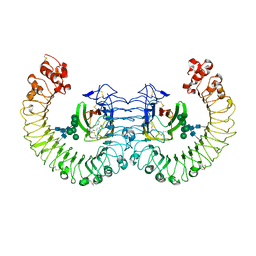 | | Crystal structure of the RP105/MD-1 complex | | Descriptor: | (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, CD180 molecule, LY86 protein, ... | | Authors: | Yoon, S.I, Hong, M, Wilson, I.A. | | Deposit date: | 2011-04-07 | | Release date: | 2011-08-31 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | An unusual dimeric structure and assembly for TLR4 regulator RP105-MD-1.
Nat.Struct.Mol.Biol., 18, 2011
|
|
2HVP
 
 | |
2HYT
 
 | |
2HRR
 
 | |
1JLQ
 
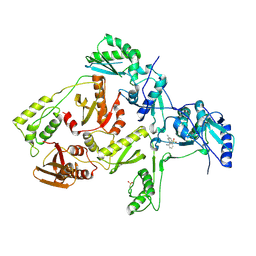 | | CRYSTAL STRUCTURE OF HIV-1 REVERSE TRANSCRIPTASE IN COMPLEX WITH 739W94 | | Descriptor: | 2-AMINO-6-(3,5-DIMETHYLPHENYL)SULFONYLBENZONITRILE, HIV-1 RT, A-CHAIN, ... | | Authors: | Ren, J, Stuart, D.I, Stammers, D.K. | | Deposit date: | 2001-07-16 | | Release date: | 2001-08-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | 2-Amino-6-arylsulfonylbenzonitriles as non-nucleoside reverse transcriptase inhibitors of HIV-1.
J.Med.Chem., 44, 2001
|
|
2F3D
 
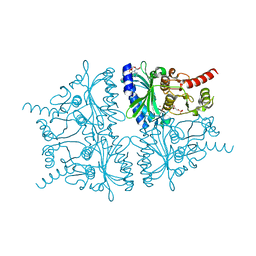 | | Mechanism of displacement of a catalytically essential loop from the active site of fructose-1,6-bisphosphatase | | Descriptor: | 6-O-phosphono-beta-D-fructofuranose, ADENOSINE MONOPHOSPHATE, Fructose-1,6-bisphosphatase 1, ... | | Authors: | Iancu, C.V, Mukund, S, Choe, J.-Y, Fromm, H.J, Honzatko, R.B. | | Deposit date: | 2005-11-21 | | Release date: | 2006-04-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Mechanism of displacement of a catalytically essential loop from the active site of mammalian fructose-1,6-bisphosphatase.
Biochemistry, 52, 2013
|
|
1B2Y
 
 | |
2BGN
 
 | | HIV-1 Tat protein derived N-terminal nonapeptide Trp2-Tat(1-9) bound to the active site of Dipeptidyl peptidase IV (CD26) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Weihofen, W.A, Liu, J, Reutter, W, Saenger, W, Fan, H. | | Deposit date: | 2005-01-03 | | Release date: | 2005-01-27 | | Last modified: | 2025-10-01 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Crystal Structures of HIV-1 Tat-Derived Nonapeptides Tat-(1-9) and Trp2-Tat-(1-9) Bound to the Active Site of Dipeptidyl-Peptidase Iv (Cd26)
J.Biol.Chem., 280, 2005
|
|
2L4L
 
 | | Structural insights into the cTAR DNA recognition by the HIV-1 Nucleocapsid protein: role of sugar deoxyriboses in the binding polarity of NC | | Descriptor: | 5'-D(*CP*TP*GP*G)-3', HIV-1 nucleocapsid protein NCp7, ZINC ION | | Authors: | Bazzi, A, Zargarian, L, Chaminade, F, Boudier, C, De Rocquigny, H, Rene, B, Mely, Y, Fosse, P, Mauffret, O. | | Deposit date: | 2010-10-08 | | Release date: | 2010-12-08 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural insights into the cTAR DNA recognition by the HIV-1 nucleocapsid protein: role of sugar deoxyriboses in the binding polarity of NC.
Nucleic Acids Res., 39, 2011
|
|
1AXK
 
 | | ENGINEERED BACILLUS BIFUNCTIONAL ENZYME GLUXYN-1 | | Descriptor: | CALCIUM ION, GLUXYN-1 | | Authors: | Ay, J, Heinemann, U. | | Deposit date: | 1997-10-16 | | Release date: | 1999-05-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and function of the Bacillus hybrid enzyme GluXyn-1: native-like jellyroll fold preserved after insertion of autonomous globular domain.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
1JLC
 
 | | CRYSTAL STRUCTURE OF Y181C MUTANT HIV-1 REVERSE TRANSCRIPTASE IN COMPLEX WITH PETT-2 | | Descriptor: | HIV-1 RT A-chain, HIV-1 RT B-chain, N-[[3-FLUORO-4-ETHOXY-PYRID-2-YL]ETHYL]-N'-[5-CHLORO-PYRIDYL]-THIOUREA | | Authors: | Ren, J, Nichols, C, Bird, L, Chamberlain, P, Weaver, K, Short, S, Stuart, D.I, Stammers, D.K. | | Deposit date: | 2001-07-16 | | Release date: | 2001-10-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural mechanisms of drug resistance for mutations at codons 181 and 188 in HIV-1 reverse transcriptase and the improved resilience of second generation non-nucleoside inhibitors.
J.Mol.Biol., 312, 2001
|
|
7KMD
 
 | |
2CYG
 
 | | Crystal structure at 1.45- resolution of the major allergen endo-beta-1,3-glucanase of banana as a molecular basis for the latex-fruit syndrome | | Descriptor: | beta-1, 3-glucananse | | Authors: | Receveur-Brechot, V, Czjzek, M, Barre, A, Roussel, A, Peumans, W.J, Van Damme, E.J.M, Rouge, P. | | Deposit date: | 2005-07-07 | | Release date: | 2005-11-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal structure at 1.45-A resolution of the major allergen endo-beta-1,3-glucanase of banana as a molecular basis for the latex-fruit syndrome
Proteins, 63, 2006
|
|
2BGR
 
 | | Crystal structure of HIV-1 Tat derived nonapeptides Tat(1-9) bound to the active site of Dipeptidyl peptidase IV (CD26) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Weihofen, W.A, Liu, J, Reutter, W, Saenger, W, Fan, H. | | Deposit date: | 2005-01-04 | | Release date: | 2005-01-27 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures of HIV-1 Tat-Derived Nonapeptides Tat-(1-9) and Trp2-Tat-(1-9) Bound to the Active Site of Dipeptidyl-Peptidase Iv (Cd26)
J.Biol.Chem., 280, 2005
|
|
1JLE
 
 | | CRYSTAL STRUCTURE OF Y188C MUTANT HIV-1 REVERSE TRANSCRIPTASE | | Descriptor: | HIV-1 RT, A-CHAIN, B-CHAIN | | Authors: | Ren, J, Nichols, C, Bird, L, Chamberlain, P, Weaver, K, Short, S, Stuart, D.I, Stammers, D.K. | | Deposit date: | 2001-07-16 | | Release date: | 2001-10-03 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural mechanisms of drug resistance for mutations at codons 181 and 188 in HIV-1 reverse transcriptase and the improved resilience of second generation non-nucleoside inhibitors.
J.Mol.Biol., 312, 2001
|
|
2GFT
 
 | | Crystal structure of the E263A nucleophile mutant of Bacillus licheniformis endo-beta-1,4-galactanase in complex with galactotriose | | Descriptor: | CALCIUM ION, Glycosyl Hydrolase Family 53, beta-D-galactopyranose-(1-4)-beta-D-galactopyranose-(1-4)-beta-D-galactopyranose | | Authors: | Welner, D, Le Nours, J, De Maria, L, Jorgensen, C.T, Christensen, L.L.H, Larsen, S, Lo Leggio, L. | | Deposit date: | 2006-03-23 | | Release date: | 2006-06-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Oligosaccharide binding of Bacillus licheniformis endo-beta-1,4-galactanase
To be Published
|
|
1JLG
 
 | | CRYSTAL STRUCTURE OF Y188C MUTANT HIV-1 REVERSE TRANSCRIPTASE IN COMPLEX WITH UC-781 | | Descriptor: | 2-METHYL-FURAN-3-CARBOTHIOIC ACID [4-CHLORO-3-(3-METHYL-BUT-2-ENYLOXY)-PHENYL]-AMIDE, HIV-1 RT A-chain, HIV-1 RT B-chain | | Authors: | Ren, J, Nichols, C, Bird, L, Chamberlain, P, Weaver, K, Short, S, Stuart, D.I, Stammers, D.K. | | Deposit date: | 2001-07-16 | | Release date: | 2001-10-03 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural mechanisms of drug resistance for mutations at codons 181 and 188 in HIV-1 reverse transcriptase and the improved resilience of second generation non-nucleoside inhibitors.
J.Mol.Biol., 312, 2001
|
|
2LAV
 
 | | NMR solution structure of human Vaccinia-Related Kinase 1 | | Descriptor: | Vaccinia-related kinase 1 | | Authors: | Shin, J, Yoon, H.S. | | Deposit date: | 2011-03-21 | | Release date: | 2011-05-04 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR Solution Structure of Human Vaccinia-related Kinase 1 (VRK1) Reveals the C-terminal Tail Essential for Its Structural Stability and Autocatalytic Activity.
J.Biol.Chem., 286, 2011
|
|
2E9N
 
 | | Structure of h-CHK1 complexed with A767085 | | Descriptor: | 3-(4'-HYDROXYBIPHENYL-4-YL)-N-(4-HYDROXYCYCLOHEXYL)-1,4-DIHYDROINDENO[1,2-C]PYRAZOLE-6-CARBOXAMIDE, Serine/threonine-protein kinase Chk1 | | Authors: | Park, C. | | Deposit date: | 2007-01-26 | | Release date: | 2008-01-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery of 1,4-dihydroindeno[1,2-c]pyrazoles as a novel class of potent and selective checkpoint kinase 1 inhibitors.
Bioorg.Med.Chem., 15, 2007
|
|
4ACU
 
 | | Aminoimidazoles as BACE-1 Inhibitors. X-RAY CRYSTAL STRUCTURE OF BETA SECRETASE COMPLEXED WITH COMPOUND 14 | | Descriptor: | (8S)-3,3-DIFLUORO-8-(2'-FLUORO-3'-METHOXYBIPHENYL-3-YL)-8-PYRIDIN-4-YL-2,3,4,8-TETRAHYDROIMIDAZO[1,5-A]PYRIMIDIN-6-AMINE, ACETATE ION, BETA-SECRETASE 1 | | Authors: | Swahn, B, Holenz, J, Kihlstrom, J, Kolmodin, K, Lindstrom, J, Plobeck, N, Rotticci, D, Sehgelmeble, F, Sundstrom, M, von Berg, S, Falting, J, Georgievska, B, Gustavsson, S, Neelissen, J, Ek, M, Olsson, L.L, Berg, S. | | Deposit date: | 2011-12-19 | | Release date: | 2012-02-29 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Aminoimidazoles as BACE-1 inhibitors: the challenge to achieve in vivo brain efficacy.
Bioorg. Med. Chem. Lett., 22, 2012
|
|
2GTZ
 
 | |
1RP8
 
 | | Crystal structure of barley alpha-amylase isozyme 1 (amy1) inactive mutant d180a in complex with maltoheptaose | | Descriptor: | Alpha-amylase type 1 isozyme, CALCIUM ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, ... | | Authors: | Robert, X, Haser, R, Aghajari, N. | | Deposit date: | 2003-12-03 | | Release date: | 2005-06-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Oligosaccharide Binding to Barley {alpha}-Amylase 1
J.Biol.Chem., 280, 2005
|
|
3GXT
 
 | |
