1XJI
 
 | | Bacteriorhodopsin crystallized in bicelles at room temperature | | Descriptor: | 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, Bacteriorhodopsin, DECANE, ... | | Authors: | Faham, S, Boulting, G.L, Massey, E.A, Yohannan, S, Yang, D, Bowie, J.U. | | Deposit date: | 2004-09-23 | | Release date: | 2005-04-19 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystallization of bacteriorhodopsin from bicelle formulations at room temperature
Protein Sci., 14, 2005
|
|
1YKR
 
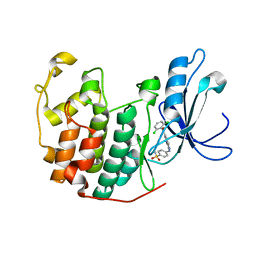 | | Crystal structure of cdk2 with an aminoimidazo pyridine inhibitor | | Descriptor: | 4-{[6-(2,6-DICHLOROBENZOYL)IMIDAZO[1,2-A]PYRIDIN-2-YL]AMINO}BENZENESULFONAMIDE, Cell division protein kinase 2 | | Authors: | Hamdouchi, C, Zhong, B, Mendoza, J, Jaramillo, C, Zhang, F, Brooks, H.B. | | Deposit date: | 2005-01-18 | | Release date: | 2006-01-24 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-based design of a new class of highly selective aminoimidazo[1,2-a]pyridine-based inhibitors of cyclin dependent kinases
Bioorg.Med.Chem.Lett., 15, 2005
|
|
2NQI
 
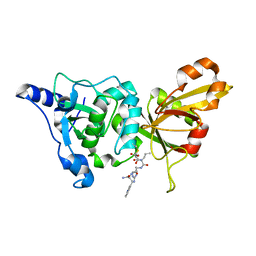 | | Calpain 1 proteolytic core inactivated by WR13(R,R), an epoxysuccinyl-type inhibitor. | | Descriptor: | CALCIUM ION, Calpain-1 catalytic subunit, N~2~-[(2S)-2-{[(2R)-4-ETHOXY-2-HYDROXY-4-OXOBUTANOYL]AMINO}PENT-4-ENOYL]-L-ARGINYL-L-TRYPTOPHANAMIDE | | Authors: | Cuerrier, D, Davies, P.L, Campbell, R.L, Moldoveanu, T. | | Deposit date: | 2006-10-31 | | Release date: | 2007-01-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Development of Calpain-specific Inactivators by Screening of Positional Scanning Epoxide Libraries
J.Biol.Chem., 282, 2007
|
|
1YMZ
 
 | | CC45, An Artificial WW Domain Designed Using Statistical Coupling Analysis | | Descriptor: | CC45 | | Authors: | Socolich, M, Lockless, S.W, Russ, W.P, Lee, H, Gardner, K.H, Ranganathan, R. | | Deposit date: | 2005-01-22 | | Release date: | 2005-09-27 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Evolutionary information for specifying a protein fold.
Nature, 437, 2005
|
|
2MVD
 
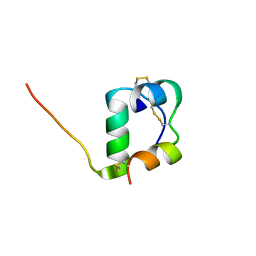 | | Solution structure of [GlnB22]-insulin mutant at pH 1.9 | | Descriptor: | Insulin A chain, Insulin B chain | | Authors: | Hexnerova, R, Krizkova, K, Maletinska, L, Jiracek, J, Brzozowski, A.M, Zakova, L, Veverka, V. | | Deposit date: | 2014-10-02 | | Release date: | 2014-12-10 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structural and Functional Study of the GlnB22-Insulin Mutant Responsible for Maturity-Onset Diabetes of the Young.
Plos One, 9, 2014
|
|
2MSA
 
 | |
3N9W
 
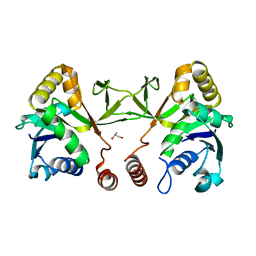 | | Crystal structure of 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase (IspD) in complex with 1,2-Propanediol | | Descriptor: | 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase, R-1,2-PROPANEDIOL, S-1,2-PROPANEDIOL | | Authors: | Behnen, J, Heine, A, Klebe, G. | | Deposit date: | 2010-05-31 | | Release date: | 2011-05-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Experimental and computational active site mapping as a starting point to fragment-based lead discovery.
Chemmedchem, 7, 2012
|
|
2MU7
 
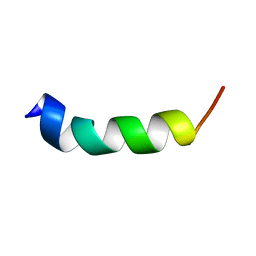 | | Shortening and modifying the 1513 MSP-1 peptide's alpha-helical region induces protection against malaria | | Descriptor: | 1513 MSP-1 peptide | | Authors: | Espejo, F, Bermudez, A, Torres, E, Urquiza, M, Rodriguez, R, Lopez, Y, Patarroyo, M. | | Deposit date: | 2014-09-04 | | Release date: | 2014-11-12 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Shortening and modifying the 1513 MSP-1 peptide's alpha-helical region induces protection against malaria.
Biochem.Biophys.Res.Commun., 315, 2004
|
|
2MSF
 
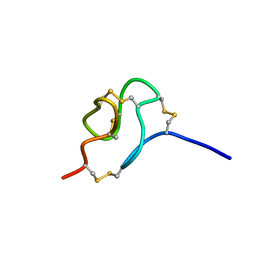 | | NMR SOLUTION STRUCTURE OF SCORPION VENOM TOXIN Ts11 (TsPep1) FROM Tityus serrulatus | | Descriptor: | Peptide TsPep1 | | Authors: | Maiti, M, Lescrinier, E, Herdewijn, P, Cremonez, C.M, Peigneur, S, Cassoli, J.S, Dutra, A.A.A, Waelkens, E, Pimenta, A.M.C, De Lima, M.H, Tytgat, J, Arantes, E.C. | | Deposit date: | 2014-08-01 | | Release date: | 2015-08-26 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structural and Functional Elucidation of Peptide Ts11 Shows Evidence of a Novel Subfamily of Scorpion Venom Toxins.
Toxins (Basel), 8, 2016
|
|
2OK4
 
 | |
2OJY
 
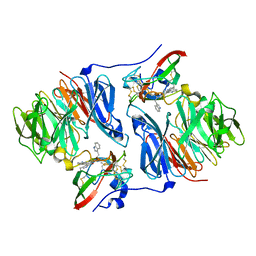 | |
2O99
 
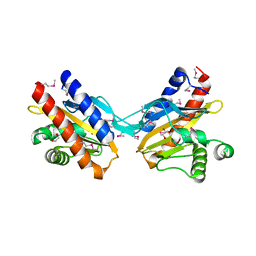 | | The crystal structure of E.coli IclR C-terminal fragment in complex with glyoxylate | | Descriptor: | 1,2-ETHANEDIOL, Acetate operon repressor, GLYCOLIC ACID | | Authors: | Lunin, V.V, Ezersky, A, Evdokimova, E, Kudritska, M, Savchenko, A. | | Deposit date: | 2006-12-13 | | Release date: | 2007-04-10 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Glyoxylate and Pyruvate Are Antagonistic Effectors of the Escherichia coli IclR Transcriptional Regulator.
J.Biol.Chem., 282, 2007
|
|
2P5G
 
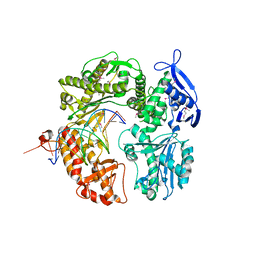 | | Crystal structure of RB69 gp43 in complex with DNA with dAMP opposite an abasic site analog in a 21mer template | | Descriptor: | DNA polymerase, Primer DNA, Template DNA | | Authors: | Zahn, K.E, Belrhali, H, Wallace, S.S, Doublie, S. | | Deposit date: | 2007-03-15 | | Release date: | 2007-10-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Caught bending the a-rule: crystal structures of translesion DNA synthesis with a non-natural nucleotide.
Biochemistry, 46, 2007
|
|
2OBG
 
 | |
3M1P
 
 | |
3MSF
 
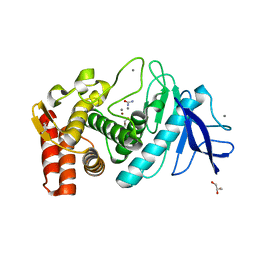 | | Crystal structure of Thermolysin in complex with Urea | | Descriptor: | CALCIUM ION, GLYCEROL, Thermolysin, ... | | Authors: | Behnen, J, Heine, A, Klebe, G. | | Deposit date: | 2010-04-29 | | Release date: | 2011-05-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Experimental and computational active site mapping as a starting point to fragment-based lead discovery.
Chemmedchem, 7, 2012
|
|
3M2N
 
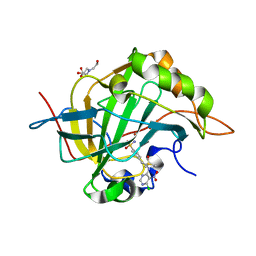 | | Crystal structure of human carbonic anhydrase isozyme II with 4-{2-[N-(6-chloro-5-nitropyrimidin-4-yl)amino]ethyl}benzenesulfonamide | | Descriptor: | 4-{2-[(6-chloro-5-nitropyrimidin-4-yl)amino]ethyl}benzenesulfonamide, BICINE, Carbonic anhydrase 2, ... | | Authors: | Grazulis, S, Manakova, E, Golovenko, D. | | Deposit date: | 2010-03-08 | | Release date: | 2011-02-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Design of [(2-pyrimidinylthio)acetyl]benzenesulfonamides as inhibitors of human carbonic anhydrases.
Eur.J.Med.Chem., 51, 2012
|
|
2O3C
 
 | | Crystal structure of zebrafish Ape | | Descriptor: | APEX nuclease 1, LEAD (II) ION | | Authors: | Georgiadis, M.M, Gaur, R.K, Delaplane, S, Svenson, J. | | Deposit date: | 2006-12-01 | | Release date: | 2007-12-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Evolution of the redox function in mammalian apurinic/apyrimidinic endonuclease
Mutat.Res., 643, 2008
|
|
2O9A
 
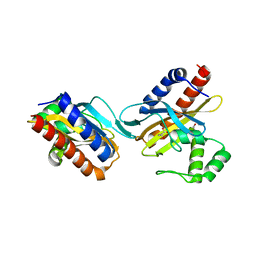 | | The crystal structure of the E.coli IclR C-terminal fragment in complex with pyruvate. | | Descriptor: | 1,2-ETHANEDIOL, Acetate operon repressor, PYRUVIC ACID | | Authors: | Lunin, V.V, Ezersky, A, Evdokimova, E, Kudritska, M, Savchenko, A. | | Deposit date: | 2006-12-13 | | Release date: | 2007-04-10 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Glyoxylate and Pyruvate Are Antagonistic Effectors of the Escherichia coli IclR Transcriptional Regulator.
J.Biol.Chem., 282, 2007
|
|
2O3H
 
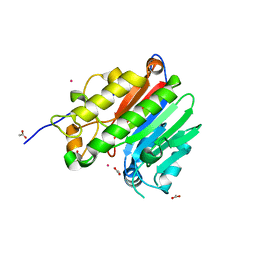 | | Crystal structure of the human C65A Ape | | Descriptor: | ACETATE ION, DNA-(apurinic or apyrimidinic site) lyase, SAMARIUM (III) ION | | Authors: | Georgiadis, M.M, Gaur, R.K, Delaplane, S, Svenson, J. | | Deposit date: | 2006-12-01 | | Release date: | 2007-12-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Evolution of the redox function in mammalian apurinic/apyrimidinic endonuclease
Mutat.Res., 643, 2008
|
|
3PD6
 
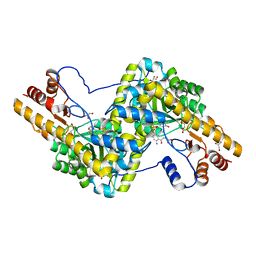 | | Crystal structure of mouse mitochondrial aspartate aminotransferase, a newly identified kynurenine aminotransferase-IV | | Descriptor: | (2S)-2-amino-4-(2-aminophenyl)-4-oxobutanoic acid, 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, Aspartate aminotransferase, ... | | Authors: | Han, Q, Robinson, H, Cai, T, Tagle, D.A, Li, J. | | Deposit date: | 2010-10-22 | | Release date: | 2010-11-10 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Biochemical and structural characterization of mouse mitochondrial aspartate aminotransferase, a newly identified kynurenine aminotransferase-IV.
Biosci.Rep., 31, 2011
|
|
3PCZ
 
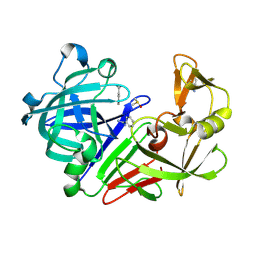 | | Endothiapepsin in complex with benzamidine | | Descriptor: | BENZAMIDINE, DIMETHYL SULFOXIDE, Endothiapepsin | | Authors: | Koester, H, Heine, A, Klebe, G. | | Deposit date: | 2010-10-22 | | Release date: | 2011-10-19 | | Last modified: | 2021-08-04 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Experimental and computational active site mapping as a starting point to fragment-based lead discovery.
Chemmedchem, 7, 2012
|
|
3PDB
 
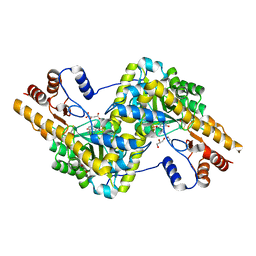 | | Crystal structure of mouse mitochondrial aspartate aminotransferase in complex with oxaloacetic acid | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, Aspartate aminotransferase, mitochondrial, ... | | Authors: | Han, Q, Robinson, H, Cai, T, Tagle, D.A, Li, J. | | Deposit date: | 2010-10-22 | | Release date: | 2010-11-10 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Biochemical and structural characterization of mouse mitochondrial aspartate aminotransferase, a newly identified kynurenine aminotransferase-IV.
Biosci.Rep., 31, 2011
|
|
2O81
 
 | |
2O83
 
 | |
