6W3L
 
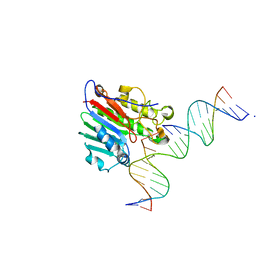 | | APE1 exonuclease substrate complex wild-type | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, DNA-(apurinic or apyrimidinic site) lyase, ... | | Authors: | Freudenthal, B.D, Whitaker, A.M. | | Deposit date: | 2020-03-09 | | Release date: | 2020-06-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Molecular and structural characterization of disease-associated APE1 polymorphisms.
DNA Repair (Amst.), 91-92, 2020
|
|
5J3G
 
 | |
5J3F
 
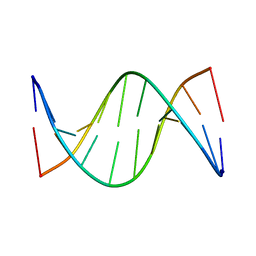 | | NMR solution structure of [Rp, Rp]-PT dsDNA | | Descriptor: | DNA (5'-D(*CP*GP*(RSG)P*CP*CP*GP*CP*CP*GP*A)-3'), DNA (5'-D(*TP*CP*GP*GP*CP*GP*(RSG)P*CP*CP*G)-3') | | Authors: | Lan, W, Hu, Z, Cao, C. | | Deposit date: | 2016-03-30 | | Release date: | 2016-11-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural investigation into physiological DNA phosphorothioate modification
Sci Rep, 6, 2016
|
|
5J3I
 
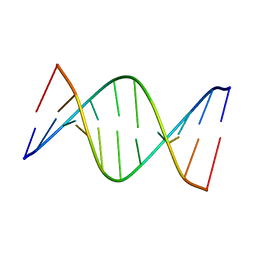 | | NMR solution structure of [Sp, Sp]-PT dsDNA | | Descriptor: | DNA (5'-D(*CP*GP*(SSG)P*CP*CP*GP*CP*CP*GP*A)-3'), DNA (5'-D(*TP*CP*GP*GP*CP*GP*(SSG)P*CP*CP*G)-3') | | Authors: | Lan, W, Hu, Z, Cao, C. | | Deposit date: | 2016-03-30 | | Release date: | 2016-11-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural investigation into physiological DNA phosphorothioate modification
Sci Rep, 6, 2016
|
|
8VXY
 
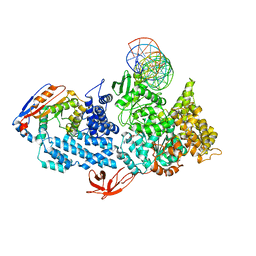 | | Structure of HamA(E138A,K140A)B-plasmid DNA complex from the Escherichia coli Hachiman defense system | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, HamA, HamB, ... | | Authors: | Tuck, O.T, Hu, J.J, Doudna, J.A. | | Deposit date: | 2024-02-06 | | Release date: | 2024-03-13 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.19 Å) | | Cite: | Hachiman is a genome integrity sensor.
Biorxiv, 2024
|
|
8W0A
 
 | |
4M9V
 
 | | Zfp57 mutant (E182Q) in complex with 5-carboxylcytosine DNA | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Liu, Y, Olanrewaju, Y.O, Zhang, X, Cheng, X. | | Deposit date: | 2013-08-15 | | Release date: | 2013-11-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (0.969 Å) | | Cite: | DNA recognition of 5-carboxylcytosine by a zfp57 mutant at an atomic resolution of 0.97 angstrom.
Biochemistry, 52, 2013
|
|
6FWR
 
 | | Structure of DinG in complex with ssDNA | | Descriptor: | ATP-dependent DNA helicase DinG, DNA (5'-D(*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*T)-3'), IRON/SULFUR CLUSTER | | Authors: | Cheng, K, Wigley, D.B. | | Deposit date: | 2018-03-07 | | Release date: | 2018-12-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | DNA translocation mechanism of an XPD family helicase.
Elife, 7, 2018
|
|
1ZQW
 
 | |
1ZQU
 
 | |
1ZQZ
 
 | |
3PVP
 
 | | Structure of Mycobacterium tuberculosis DnaA-DBD in complex with box2 DNA | | Descriptor: | Chromosomal replication initiator protein dnaA, DNA (5'-D(*CP*GP*TP*TP*AP*TP*CP*CP*AP*CP*AP*AP*C)-3'), DNA (5'-D(*GP*TP*TP*GP*TP*GP*GP*AP*TP*AP*AP*CP*G)-3') | | Authors: | Tsodikov, O.V, Biswas, T. | | Deposit date: | 2010-12-07 | | Release date: | 2011-05-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and Thermodynamic Signatures of DNA Recognition by Mycobacterium tuberculosis DnaA.
J.Mol.Biol., 410, 2011
|
|
1ZQY
 
 | |
1ZQX
 
 | |
6FWS
 
 | | Structure of DinG in complex with ssDNA and ADPBeF | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent DNA helicase DinG, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Cheng, K, Wigley, D. | | Deposit date: | 2018-03-07 | | Release date: | 2018-12-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | DNA translocation mechanism of an XPD family helicase.
Elife, 7, 2018
|
|
221D
 
 | |
7LWA
 
 | |
7LW9
 
 | | Human Exonuclease 5 crystal structure in complex with ssDNA, Sm, and Na | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, DNA (5'-D(*AP*TP*TP*GP*CP*TP*GP*AP*AP*GP*GP*G)-3'), ... | | Authors: | Tsai, C.L, Tainer, J.A. | | Deposit date: | 2021-02-28 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | EXO5-DNA structure and BLM interactions direct DNA resection critical for ATR-dependent replication restart.
Mol.Cell, 81, 2021
|
|
3PVV
 
 | | Structure of Mycobacterium tuberculosis DnaA-DBD in complex with box1 DNA | | Descriptor: | Chromosomal replication initiator protein dnaA, DNA (5'-D(*CP*GP*TP*TP*GP*TP*CP*CP*AP*CP*AP*AP*C)-3'), DNA (5'-D(*GP*TP*TP*GP*TP*GP*GP*AP*CP*AP*AP*CP*G)-3') | | Authors: | Tsodikov, O.V, Biswas, T. | | Deposit date: | 2010-12-07 | | Release date: | 2011-05-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and Thermodynamic Signatures of DNA Recognition by Mycobacterium tuberculosis DnaA.
J.Mol.Biol., 410, 2011
|
|
7LW8
 
 | | Human Exonuclease 5 crystal structure in complex with a ssDNA | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*T)-3'), Exonuclease V, ... | | Authors: | Tsai, C.L, Tainer, J.A. | | Deposit date: | 2021-02-28 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | EXO5-DNA structure and BLM interactions direct DNA resection critical for ATR-dependent replication restart.
Mol.Cell, 81, 2021
|
|
7BHY
 
 | | DNA-binding domain of DeoR in complex with the DNA operator | | Descriptor: | DNA operator - strand 1, DNA operator - strand 2, Deoxyribonucleoside regulator, ... | | Authors: | Novakova, M, Rezacova, P, Skerlova, J, Brynda, J. | | Deposit date: | 2021-01-11 | | Release date: | 2021-11-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural insight into DNA recognition by bacterial transcriptional regulators of the SorC/DeoR family.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
8DC2
 
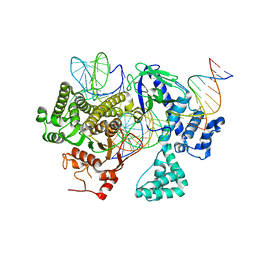 | | Cryo-EM structure of CasLambda (Cas12l) bound to crRNA and DNA | | Descriptor: | CasLambda, DNA NTS, DNA TS, ... | | Authors: | Al-Shayeb, B, Skopintsev, P, Soczek, K, Doudna, J. | | Deposit date: | 2022-06-15 | | Release date: | 2022-12-14 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.99 Å) | | Cite: | Diverse virus-encoded CRISPR-Cas systems include streamlined genome editors.
Cell, 185, 2022
|
|
4LYL
 
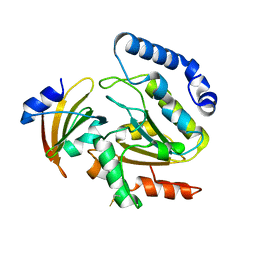 | | Crystal structure of uracil-DNA glycosylase from cod (Gadus morhua) in complex with the proteinaceous inhibitor UGI | | Descriptor: | Uracil-DNA glycosylase, Uracil-DNA glycosylase inhibitor | | Authors: | Assefa, N.G, Niiranen, L.M.K, Johnson, K.A, Leiros, H.-K.S, Smalas, A.O, Willassen, N.P, Moe, E. | | Deposit date: | 2013-07-31 | | Release date: | 2014-08-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structural and biophysical analysis of interactions between cod and human uracil-DNA N-glycosylase (UNG) and UNG inhibitor (Ugi).
Acta Crystallogr.,Sect.D, 70, 2014
|
|
8E8A
 
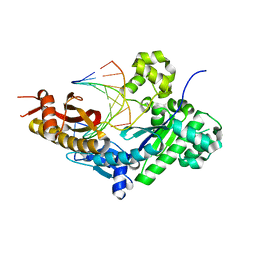 | | Human DNA polymerase eta-DNA-dT-ended primer binary complex | | Descriptor: | DNA (5'-D(*AP*GP*CP*GP*TP*CP*AP*T)-3'), DNA (5'-D(*CP*AP*TP*TP*AP*TP*GP*AP*CP*GP*CP*T)-3'), DNA polymerase eta | | Authors: | Chang, C, Gao, Y. | | Deposit date: | 2022-08-25 | | Release date: | 2023-02-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Primer terminal ribonucleotide alters the active site dynamics of DNA polymerase eta and reduces DNA synthesis fidelity.
J.Biol.Chem., 299, 2023
|
|
8E89
 
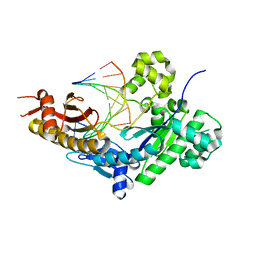 | | Human DNA polymerase eta-DNA-rU-ended primer-binary complex | | Descriptor: | DNA (5'-D(*CP*AP*TP*TP*AP*TP*GP*AP*CP*GP*CP*T)-3'), DNA polymerase eta, DNA/RNA (5'-D(*AP*GP*CP*GP*TP*CP*A)-R(P*U)-3') | | Authors: | Chang, C, Gao, Y. | | Deposit date: | 2022-08-25 | | Release date: | 2023-02-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Primer terminal ribonucleotide alters the active site dynamics of DNA polymerase eta and reduces DNA synthesis fidelity.
J.Biol.Chem., 299, 2023
|
|
