5QQM
 
 | | PanDDA analysis group deposition -- Crystal Structure of Kalirin/Rac1 in complex with MolPort-009-565-325 | | Descriptor: | 1,2-ETHANEDIOL, Kalirin, N-(1-benzyl-1H-pyrazol-4-yl)-N'-(5-methyl-1,2-oxazol-3-yl)urea, ... | | Authors: | Gray, J.L, Krojer, T, Talon, R, Douangamath, A, Jimenez Antunez, C, Bountra, C, Arrowsmith, C.H, Edwards, A, Brennan, P.E, von Delft, F. | | Deposit date: | 2019-05-03 | | Release date: | 2019-12-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5QQF
 
 | | PanDDA analysis group deposition -- Crystal Structure of Kalirin/Rac1 in complex with MolPort-004-412-710 | | Descriptor: | 1,2-ETHANEDIOL, Kalirin, N-ethyl-N'-1,3-thiazol-2-ylurea, ... | | Authors: | Gray, J.L, Krojer, T, Talon, R, Douangamath, A, Jimenez Antunez, C, Bountra, C, Arrowsmith, C.H, Edwards, A, Brennan, P.E, von Delft, F. | | Deposit date: | 2019-05-03 | | Release date: | 2019-12-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5QQJ
 
 | | PanDDA analysis group deposition -- Crystal Structure of Kalirin/Rac1 in complex with MolPort-009-587-558 | | Descriptor: | 1,2-ETHANEDIOL, Kalirin, N-{1-[(imidazo[1,2-a]pyridin-2-yl)methyl]-1H-pyrazol-4-yl}-N'-(5-methyl-1,2-oxazol-3-yl)urea, ... | | Authors: | Gray, J.L, Krojer, T, Talon, R, Douangamath, A, Jimenez Antunez, C, Bountra, C, Arrowsmith, C.H, Edwards, A, Brennan, P.E, von Delft, F. | | Deposit date: | 2019-05-03 | | Release date: | 2019-12-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5QQL
 
 | | PanDDA analysis group deposition -- Crystal Structure of Kalirin/Rac1 in complex with MolPort-009-565-301 | | Descriptor: | 1,2-ETHANEDIOL, Kalirin, N-(5-methyl-1,2-oxazol-3-yl)-N'-[2-(phenylsulfonyl)ethyl]urea, ... | | Authors: | Gray, J.L, Krojer, T, Talon, R, Douangamath, A, Jimenez Antunez, C, Bountra, C, Arrowsmith, C.H, Edwards, A, Brennan, P.E, von Delft, F. | | Deposit date: | 2019-05-03 | | Release date: | 2019-12-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5K5F
 
 | | NMR structure of the HLTF HIRAN domain | | Descriptor: | Helicase-like transcription factor | | Authors: | Bezsonova, I, Neculai, D, Korzhnev, D, Weigelt, J, Bountra, C, Edwards, A, Arrowsmith, C, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-05-23 | | Release date: | 2016-06-08 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the HLTF HIRAN domain
To Be Published
|
|
5QQH
 
 | | PanDDA analysis group deposition -- Crystal Structure of Kalirin/Rac1 in complex with MolPort-020-096-465 | | Descriptor: | 1,2-ETHANEDIOL, Kalirin, N-methyl-5-({[(5-methyl-1,2-oxazol-3-yl)carbamoyl]amino}methyl)thiophene-2-sulfonamide, ... | | Authors: | Gray, J.L, Krojer, T, Talon, R, Douangamath, A, Jimenez Antunez, C, Bountra, C, Arrowsmith, C.H, Edwards, A, Brennan, P.E, von Delft, F. | | Deposit date: | 2019-05-03 | | Release date: | 2019-12-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5QQI
 
 | | PanDDA analysis group deposition -- Crystal Structure of Kalirin/Rac1 in complex with MolPort-009-178-994 | | Descriptor: | 1,2-ETHANEDIOL, Kalirin, N-[3-(methoxymethyl)phenyl]-N'-(5-methyl-1,2-oxazol-3-yl)urea, ... | | Authors: | Gray, J.L, Krojer, T, Talon, R, Douangamath, A, Jimenez Antunez, C, Bountra, C, Arrowsmith, C.H, Edwards, A, Brennan, P.E, von Delft, F. | | Deposit date: | 2019-05-03 | | Release date: | 2019-12-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5QU9
 
 | | PanDDA analysis group deposition of ground-state model of Kalirin/Rac1 screened against a customized urea fragment library by X-ray Crystallography at the XChem facility of Diamond Light Source beamline I04-1 | | Descriptor: | 1,2-ETHANEDIOL, Kalirin, Ras-related C3 botulinum toxin substrate 1 | | Authors: | Gray, J.L, Krojer, T, Talon, R, Douangamath, A, Jimenez Antunez, C, Bountra, C, Arrowsmith, C.H, Edwards, A, Brennan, P.E, von Delft, F. | | Deposit date: | 2019-12-13 | | Release date: | 2020-01-29 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | PanDDA analysis group deposition of ground-state model
To Be Published
|
|
5QQD
 
 | | PanDDA analysis group deposition -- Crystal Structure of Kalirin/Rac1 in complex with Z56880342 | | Descriptor: | 1,2-ETHANEDIOL, Kalirin, N-ethyl-N'-(5-methyl-1,2-oxazol-3-yl)urea, ... | | Authors: | Gray, J.L, Krojer, T, Talon, R, Douangamath, A, Jimenez Antunez, C, Bountra, C, Arrowsmith, C.H, Edwards, A, Brennan, P.E, von Delft, F. | | Deposit date: | 2019-05-03 | | Release date: | 2019-12-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5L0C
 
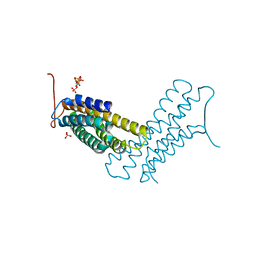 | | Human metavinculin (residues 959-1134) in complex with PIP2 | | Descriptor: | PHOSPHATE ION, Vinculin, [(2R)-2-octanoyloxy-3-[oxidanyl-[(1R,2R,3S,4R,5R,6S)-2,3,6-tris(oxidanyl)-4,5-diphosphonooxy-cyclohexyl]oxy-phosphoryl]oxy-propyl] octanoate | | Authors: | Chinthalapudi, K, Izard, T. | | Deposit date: | 2016-07-27 | | Release date: | 2016-08-31 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Differential lipid binding of vinculin isoforms promotes quasi-equivalent dimerization.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5L0F
 
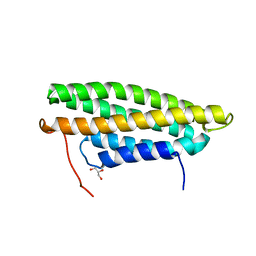 | |
5FEF
 
 | |
5L0G
 
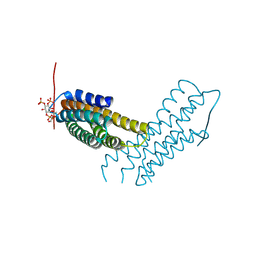 | | Human metavinculin MVt Q971R, R975D, T978R mutant (residues 959-1134) in complex with PIP2 | | Descriptor: | Vinculin, [(2R)-2-octanoyloxy-3-[oxidanyl-[(1R,2R,3S,4R,5R,6S)-2,3,6-tris(oxidanyl)-4,5-diphosphonooxy-cyclohexyl]oxy-phosphoryl]oxy-propyl] octanoate | | Authors: | Chinthalapudi, K, Izard, T. | | Deposit date: | 2016-07-27 | | Release date: | 2016-08-31 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Differential lipid binding of vinculin isoforms promotes quasi-equivalent dimerization.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5L0D
 
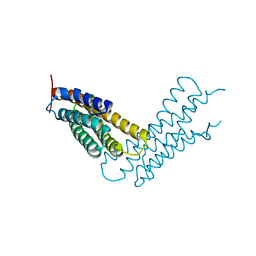 | | Human Metavinculin(residues 959-1130) in complex with PIP2 | | Descriptor: | Vinculin, [(2R)-2-octanoyloxy-3-[oxidanyl-[(1R,2R,3S,4R,5R,6S)-2,3,6-tris(oxidanyl)-4,5-diphosphonooxy-cyclohexyl]oxy-phosphoryl]oxy-propyl] octanoate | | Authors: | Chinthalapudi, K, Izard, T. | | Deposit date: | 2016-07-27 | | Release date: | 2016-08-31 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Differential lipid binding of vinculin isoforms promotes quasi-equivalent dimerization.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5L0H
 
 | |
6AJ4
 
 | |
6AJL
 
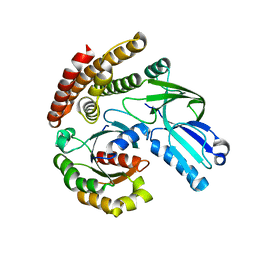 | | DOCK7 mutant I1836Y complexed with Cdc42 | | Descriptor: | Cell division control protein 42 homolog, Dedicator of cytokinesis protein 7 | | Authors: | Kukimoto-Niino, M, Shirouzu, M. | | Deposit date: | 2018-08-28 | | Release date: | 2019-03-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.23 Å) | | Cite: | Structural Basis for the Dual Substrate Specificity of DOCK7 Guanine Nucleotide Exchange Factor.
Structure, 27, 2019
|
|
2K3T
 
 | |
1E96
 
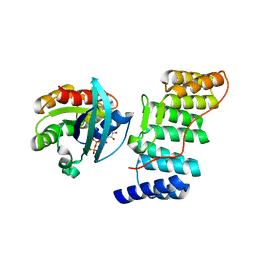 | | Structure of the Rac/p67phox complex | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, NEUTROPHIL CYTOSOL FACTOR 2 (NCF-2) TPR DOMAIN, ... | | Authors: | Lapouge, K, Smith, S.J.M, Walker, P.A, Gamblin, S.J, Smerdon, S.J, Rittinger, K. | | Deposit date: | 2000-10-10 | | Release date: | 2000-11-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the TPR domain of p67phox in complex with Rac.GTP.
Mol.Cell, 6, 2000
|
|
1T3X
 
 | |
1T3Y
 
 | |
1FOE
 
 | |
4XSH
 
 | | The complex structure of C3cer exoenzyme and GTP bound RhoA (NADH-bound state) | | Descriptor: | 1,2-ETHANEDIOL, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, ... | | Authors: | Toda, A, Tsurumura, T, Yoshida, T, Tsuge, H. | | Deposit date: | 2015-01-22 | | Release date: | 2015-06-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Rho GTPase Recognition by C3 Exoenzyme Based on C3-RhoA Complex Structure.
J.Biol.Chem., 290, 2015
|
|
4XSG
 
 | | The complex structure of C3cer exoenzyme and GTP bound RhoA (NADH-free state) | | Descriptor: | 1,2-ETHANEDIOL, 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, ADP-ribosyltransferase, ... | | Authors: | Toda, A, Tsurumura, T, Yoshida, T, Tsuge, H. | | Deposit date: | 2015-01-22 | | Release date: | 2015-06-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Rho GTPase Recognition by C3 Exoenzyme Based on C3-RhoA Complex Structure.
J.Biol.Chem., 290, 2015
|
|
4YKN
 
 | | Pi3K alpha lipid kinase with Active Site Inhibitor | | Descriptor: | 3-(6-methoxypyridin-3-yl)-5-[({4-[(5-methyl-1,3,4-thiadiazol-2-yl)sulfamoyl]phenyl}amino)methyl]benzoic acid, Phosphatidylinositol 3-kinase regulatory subunit alpha,Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform fusion protein | | Authors: | Elkins, P.A. | | Deposit date: | 2015-03-04 | | Release date: | 2015-06-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Discovery of a Potent Class of PI3K alpha Inhibitors with Unique Binding Mode via Encoded Library Technology (ELT).
Acs Med.Chem.Lett., 6, 2015
|
|
