3K5T
 
 | | Crystal structure of human diamine oxidase in space group C2221 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | McGrath, A.P, Guss, J.M. | | Deposit date: | 2009-10-08 | | Release date: | 2010-02-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | A new crystal form of human diamine oxidase.
Acta Crystallogr.,Sect.F, 66, 2010
|
|
4LW6
 
 | | Crystal structure of catalytic domain of Drosophila beta1,4galactosyltransferase 7 complex with xylobiose | | Descriptor: | 2-AMINO-1,3-PROPANEDIOL, Beta-4-galactosyltransferase 7, MANGANESE (II) ION, ... | | Authors: | Qasba, P.K, Ramakrishnan, B. | | Deposit date: | 2013-07-26 | | Release date: | 2013-09-25 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structures of beta-1,4-Galactosyltransferase 7 Enzyme Reveal Conformational Changes and Substrate Binding.
J.Biol.Chem., 288, 2013
|
|
6LVX
 
 | | Crystal structure of TLR7/Cpd-1 (SM-374527) complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 6-azanyl-2-butoxy-9-(phenylmethyl)-7H-purin-8-one, ... | | Authors: | Zhang, Z, Ohto, U, Shimizu, T. | | Deposit date: | 2020-02-06 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Structural analysis reveals TLR7 dynamics underlying antagonism.
Nat Commun, 11, 2020
|
|
6MAM
 
 | | Cleaved Ebola GP in complex with a broadly neutralizing human antibody, ADI-15946 | | Descriptor: | ADI-15946 Fab Heavy Chain, ADI-15946 Fab Light Chain, Envelope glycoprotein, ... | | Authors: | West, B.R, Moyer, C.L, Fusco, M.L, Saphire, E.O. | | Deposit date: | 2018-08-28 | | Release date: | 2019-03-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (4.1 Å) | | Cite: | Structural basis of broad ebolavirus neutralization by a human survivor antibody.
Nat. Struct. Mol. Biol., 26, 2019
|
|
5A18
 
 | |
3L4U
 
 | | Crystal complex of N-terminal Human Maltase-Glucoamylase with de-O-sulfonated kotalanol | | Descriptor: | (2R,3S,4S)-1-[(2S,3S,4R,5R,6S)-2,3,4,5,6,7-hexahydroxyheptyl]-3,4-dihydroxy-2-(hydroxymethyl)tetrahydrothiophenium (non-preferred name), 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Sim, L, Rose, D.R. | | Deposit date: | 2009-12-21 | | Release date: | 2010-02-09 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | New glucosidase inhibitors from an ayurvedic herbal treatment for type 2 diabetes: structures and inhibition of human intestinal maltase-glucoamylase with compounds from Salacia reticulata.
Biochemistry, 49, 2010
|
|
1FJ0
 
 | |
7WJ4
 
 | |
6QSW
 
 | | Complement factor B protease domain in complex with the reversible inhibitor N-(2-bromo-4-methylnaphthalen-1-yl)-4,5-dihydro-1H-imidazol-2-amine. | | Descriptor: | Complement factor B, SULFATE ION, ~{N}-(2-bromanyl-4-methyl-naphthalen-1-yl)-4,5-dihydro-1~{H}-imidazol-2-amine | | Authors: | Adams, C.M, Sellner, H, Ehara, T, Mac Sweeney, A, Crowley, M, Anderson, K, Karki, R, Mainolfi, N, Valeur, E, Sirockin, F, Gerhartz, B, Erbel, P, Hughes, N, Smith, T.M, Cumin, F, Argikar, U, Mogi, M, Sedrani, R, Wiesmann, C, Jaffee, B, Maibaum, J, Flohr, S, Harrison, R, Eder, J. | | Deposit date: | 2019-02-22 | | Release date: | 2019-03-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Small-molecule factor B inhibitor for the treatment of complement-mediated diseases.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
7WIZ
 
 | | Structural basis for ligand binding modes of CTP synthase | | Descriptor: | CTP synthase, GLUTAMINE, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, ... | | Authors: | Liu, J.L, Guo, C.J. | | Deposit date: | 2022-01-05 | | Release date: | 2023-01-11 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis for ligand binding modes of CTP synthase.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
3L4Y
 
 | | Crystal complex of N-terminal Human Maltase-Glucoamylase with NR4-8II | | Descriptor: | (1S,2R,3S,4R)-1-{(1S)-2-[(2R,3S,4S)-3,4-dihydroxy-2-(hydroxymethyl)tetrahydrothiophenium-1-yl]-1-hydroxyethyl}-2,3,4,5-tetrahydroxypentyl sulfate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Sim, L, Rose, D.R. | | Deposit date: | 2009-12-21 | | Release date: | 2010-02-09 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | New glucosidase inhibitors from an ayurvedic herbal treatment for type 2 diabetes: structures and inhibition of human intestinal maltase-glucoamylase with compounds from Salacia reticulata.
Biochemistry, 49, 2010
|
|
6QP8
 
 | | Drosophila Semaphorin 2b | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CITRATE ANION, ... | | Authors: | Robinson, R.A, Rozbesky, D, Harlos, K, Siebold, C, Jones, E.Y. | | Deposit date: | 2019-02-13 | | Release date: | 2019-08-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Diversity of oligomerization in Drosophila semaphorins suggests a mechanism of functional fine-tuning.
Nat Commun, 10, 2019
|
|
6M0U
 
 | |
6M0T
 
 | | Crystal Structure of Lysyl-tRNA Synthetase from Plasmodium falciparum complexed with L-lysine and Cladosporin derivative (CL-2) | | Descriptor: | (3R)-3-[(R)-[(2R,6S)-6-methyloxan-2-yl]-oxidanyl-methyl]-6,8-bis(oxidanyl)-3,4-dihydroisochromen-1-one, LYSINE, Lysine--tRNA ligase | | Authors: | Babbar, P, Sharma, A, Manickam, Y. | | Deposit date: | 2020-02-22 | | Release date: | 2021-04-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Design, Synthesis, and Structural Analysis of Cladosporin-Based Inhibitors of Malaria Parasites.
Acs Infect Dis., 7, 2021
|
|
6SGP
 
 | | X-ray structure of human glutamate carboxypeptidase II (GCPII) - the E424M inactive mutant, in complex with a sulfamide inhibitor GluGlu | | Descriptor: | (2~{S})-2-[[(2~{S})-1,5-bis(oxidanyl)-1,5-bis(oxidanylidene)pentan-2-yl]sulfamoylamino]pentanedioic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Barinka, C, Shukla, S, Motlova, L. | | Deposit date: | 2019-08-05 | | Release date: | 2020-08-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Structural, Biochemical, and Computational Characterization of Sulfamides as Bimetallic Peptidase Inhibitors.
J.Chem.Inf.Model., 2024
|
|
6MHA
 
 | | dHP1 Chromodomain Y24W variant bound to histone H3 peptide containing trimethyllysine | | Descriptor: | Heterochromatin protein 1, histone H3 peptide containing trimethyllysine, H3K9me3 | | Authors: | Brustad, E.M, Krone, M.W, Albanese, K.I, Waters, M.L. | | Deposit date: | 2018-09-17 | | Release date: | 2019-09-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.497 Å) | | Cite: | Thermodynamic consequences of Tyr to Trp mutations in the cation-pi-mediated binding of trimethyllysine by the HP1 chromodomain
Chem Sci, 11, 2020
|
|
6RBC
 
 | | X-ray structure of human glutamate carboxypeptidase II (GCPII) in complex with a inhibitor KB1157 | | Descriptor: | (2~{S})-2-[[(2~{R})-2-[4-[(4-iodophenyl)carbonylamino]butanoylamino]-3-oxidanyl-3-oxidanylidene-propyl]carbamoylamino]pentanedioic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Barinka, C, Kutil, Z. | | Deposit date: | 2019-04-10 | | Release date: | 2020-05-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Novel beta- and gamma-Amino Acid-Derived Inhibitors of Prostate-Specific Membrane Antigen.
J.Med.Chem., 63, 2020
|
|
3K1L
 
 | | Crystal Structure of FANCL | | Descriptor: | CITRIC ACID, Fancl, GOLD ION, ... | | Authors: | Cole, A.R, Walden, H. | | Deposit date: | 2009-09-28 | | Release date: | 2010-02-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The structure of the catalytic subunit FANCL of the Fanconi anemia core complex
Nat.Struct.Mol.Biol., 17, 2010
|
|
6MLU
 
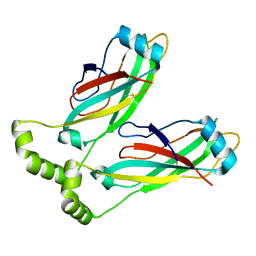 | | Cryo-EM structure of lipid droplet formation protein Seipin/BSCL2 | | Descriptor: | Seipin | | Authors: | Sui, X, Arlt, H, Liao, M, Walther, C.T, Farese, V.R. | | Deposit date: | 2018-09-28 | | Release date: | 2018-10-17 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Cryo-electron microscopy structure of the lipid droplet-formation protein seipin.
J. Cell Biol., 217, 2018
|
|
3JWD
 
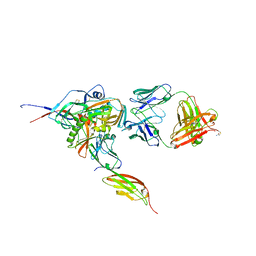 | | Structure of HIV-1 gp120 with gp41-Interactive Region: Layered Architecture and Basis of Conformational Mobility | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, FAB 48D HEAVY CHAIN, FAB 48D LIGHT CHAIN, ... | | Authors: | Pancera, M, Majeed, S, Ban, Y.A, Chen, L, Huang, C.C, Kong, L, Kwon, Y.D, Stuckey, J, Zhou, T, Robinson, J.E, Schief, W.R, Sodroski, J, Wyatt, R, Kwong, P.D. | | Deposit date: | 2009-09-18 | | Release date: | 2009-12-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Structure of HIV-1 gp120 with gp41-interactive region reveals layered envelope architecture and basis of conformational mobility.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
6RAV
 
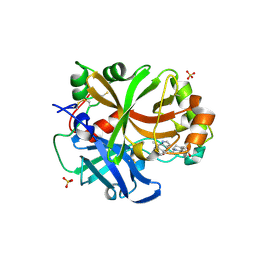 | | Complement factor B protease domain in complex with the reversible inhibitor 4-((2S,4S)-4-ethoxy-1-((5-methoxy-7-methyl-1H-indol-4-yl)methyl)piperidin-2-yl)benzoic acid | | Descriptor: | 4-[(2~{S},4~{S})-4-ethoxy-1-[(5-methoxy-7-methyl-1~{H}-indol-4-yl)methyl]piperidin-2-yl]benzoic acid, Complement factor B, SULFATE ION, ... | | Authors: | Adams, C.M, Sellner, H, Ehara, T, Mac Sweeney, A, Crowley, M, Anderson, K, Karki, R, Mainolfi, N, Valeur, E, Sirockin, F, Gerhartz, B, Erbel, P, Hughes, N, Smith, T.M, Cumin, F, Argikar, U, Mogi, M, Sedrani, R, Wiesmann, C, Jaffee, B, Maibaum, J, Flohr, S, Harrison, R, Eder, J. | | Deposit date: | 2019-04-08 | | Release date: | 2019-04-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Small-molecule factor B inhibitor for the treatment of complement-mediated diseases.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6RCU
 
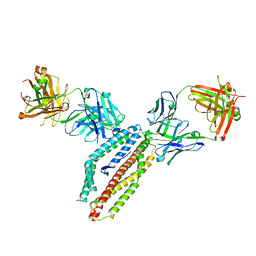 | | PfRH5 bound to monoclonal antibodies R5.004 and R5.016 | | Descriptor: | R5.004 heavy chain, R5.004 light chain, R5.016 heavy chain, ... | | Authors: | Alanine, D.W.G, Draper, S.J, Higgins, M.K. | | Deposit date: | 2019-04-11 | | Release date: | 2019-06-26 | | Last modified: | 2019-08-21 | | Method: | X-RAY DIFFRACTION (4.005 Å) | | Cite: | Human Antibodies that Slow Erythrocyte Invasion Potentiate Malaria-Neutralizing Antibodies.
Cell, 178, 2019
|
|
6SIE
 
 | | Crystal structure of the C-lobe of drosophila Arc 2 | | Descriptor: | Activity-regulated cytoskeleton associated protein 2, SULFATE ION | | Authors: | Hallin, E.I, Kursula, P. | | Deposit date: | 2019-08-09 | | Release date: | 2019-08-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal and solution structures reveal oligomerization of individual capsid homology domains of Drosophila Arc.
Plos One, 16, 2021
|
|
4NPP
 
 | | The GLIC-His10 wild-type structure in equilibrium between the open and locally-closed (LC) forms | | Descriptor: | NICKEL (II) ION, Proton-gated ion channel | | Authors: | Sauguet, L, Shahsavar, A, Poitevin, F, Huon, C, Menny, A, Nemecz, A, Haouz, A, Changeux, J.P, Corringer, P.J, Delarue, M. | | Deposit date: | 2013-11-22 | | Release date: | 2013-12-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Crystal structures of a pentameric ligand-gated ion channel provide a mechanism for activation.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4NGN
 
 | | Crystal Structure of Glutamate Carboxypeptidase II in a complex with urea-based inhibitor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Tykvart, J, Pachl, P. | | Deposit date: | 2013-11-02 | | Release date: | 2014-06-18 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Rational design of urea-based glutamate carboxypeptidase II (GCPII) inhibitors as versatile tools for specific drug targeting and delivery.
Bioorg.Med.Chem., 22, 2014
|
|
