7LU8
 
 | |
5VI6
 
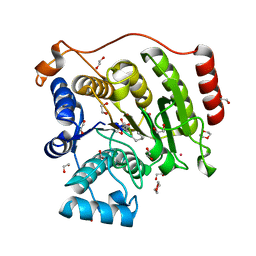 | | Crystal structure of histone deacetylase 8 in complex with trapoxin A | | Descriptor: | 1,2-ETHANEDIOL, 1,4-DIETHYLENE DIOXIDE, Histone deacetylase 8, ... | | Authors: | Porter, N.J, Christianson, D.W. | | Deposit date: | 2017-04-14 | | Release date: | 2017-09-06 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.237 Å) | | Cite: | Binding of the Microbial Cyclic Tetrapeptide Trapoxin A to the Class I Histone Deacetylase HDAC8.
ACS Chem. Biol., 12, 2017
|
|
7M0G
 
 | |
6PD5
 
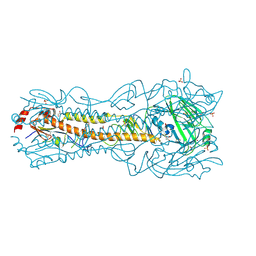 | | Crystal Structure of a H5N1 influenza virus hemagglutinin at pH 6.5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Hemagglutinin, ... | | Authors: | Antanasijevic, A, Durst, M.A, Lavie, A, Caffrey, M. | | Deposit date: | 2019-06-18 | | Release date: | 2019-11-13 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Identification of a pH sensor in Influenza hemagglutinin using X-ray crystallography.
J.Struct.Biol., 209, 2020
|
|
5IDK
 
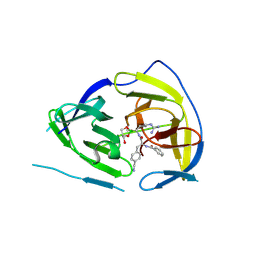 | |
7M25
 
 | |
5VIE
 
 | | Electrophilic probes for deciphering substrate recognition by O-GlcNAc transferase | | Descriptor: | 2-{[(2E)-4-chlorobut-2-enoyl]amino}-2-deoxy-beta-D-glucopyranose, 2-{[(2E)-but-2-enoyl]amino}-2-deoxy-beta-D-glucopyranose, CKII, ... | | Authors: | Jiang, J, Li, B, Hu, C.-W, Worth, M, Fan, D, Li, H. | | Deposit date: | 2017-04-15 | | Release date: | 2017-10-18 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Electrophilic probes for deciphering substrate recognition by O-GlcNAc transferase.
Nat. Chem. Biol., 13, 2017
|
|
7LL7
 
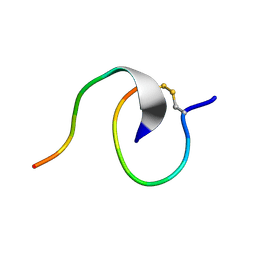 | | [2]Catenane From MccJ25 Variant G12C G21C | | Descriptor: | GLY-GLY-ALA-GLY-HIS-VAL-PRO-GLU-TYR-PHE, VAL-CYS-ILE-GLY-THR-PRO-ILE-SER-PHE-TYR-CYS | | Authors: | Link, A.J, Schroeder, H.V. | | Deposit date: | 2021-02-03 | | Release date: | 2021-08-04 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Dynamic covalent self-assembly of mechanically interlocked molecules solely made from peptides.
Nat.Chem., 13, 2021
|
|
7LUW
 
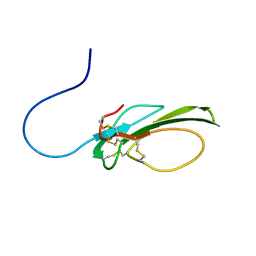 | |
5IL0
 
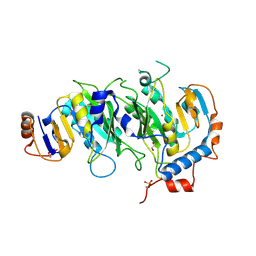 | | Crystal structural of the METTL3-METTL14 complex for N6-adenosine methylation | | Descriptor: | 1,2-ETHANEDIOL, BROMIDE ION, METTL14, ... | | Authors: | Wang, X, Guan, Z, Zou, T, Yin, P. | | Deposit date: | 2016-03-04 | | Release date: | 2016-05-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.882 Å) | | Cite: | Structural basis of N6-adenosine methylation by the METTL3-METTL14 complex
Nature, 534, 2016
|
|
7LI2
 
 | | Omega ester peptide pre-fuscimiditide | | Descriptor: | Pre-fuscimiditide peptide | | Authors: | Link, A.J, Elashal, H.E. | | Deposit date: | 2021-01-26 | | Release date: | 2022-02-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Biosynthesis and characterization of fuscimiditide, an aspartimidylated graspetide.
Nat.Chem., 14, 2022
|
|
7LNS
 
 | | NMR solution structure of PsDef2 defensin from P. sylvestris | | Descriptor: | Defensin-2 | | Authors: | Nesmelova, I.V, Kovaleva, V, Hrunyk, N.I, Yusypovych, Y.M, Shalovylo, Y.I. | | Deposit date: | 2021-02-08 | | Release date: | 2022-03-30 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Structure, dynamics, and function of PsDef2 defensin from Pinus sylvestris.
Structure, 30, 2022
|
|
5W2I
 
 | | Crystal structure of the core catalytic domain of human inositol phosphate multikinase soaked with C4-analogue of PtdIns(4,5)P2 and ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, Inositol polyphosphate multikinase,Inositol polyphosphate multikinase, ... | | Authors: | Wang, H, Shears, S.B. | | Deposit date: | 2017-06-06 | | Release date: | 2017-09-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural features of human inositol phosphate multikinase rationalize its inositol phosphate kinase and phosphoinositide 3-kinase activities.
J. Biol. Chem., 292, 2017
|
|
5IL7
 
 | |
7M1D
 
 | |
1SL6
 
 | | Crystal Structure of a fragment of DC-SIGNR (containg the carbohydrate recognition domain and two repeats of the neck) complexed with Lewis-x. | | Descriptor: | C-type lectin DC-SIGNR, CALCIUM ION, alpha-L-fucopyranose-(1-3)-[beta-D-galactopyranose-(1-4)]2-acetamido-2-deoxy-alpha-D-glucopyranose | | Authors: | Guo, Y, Feinberg, H, Conroy, E, Mitchell, D.A, Alvarez, R, Blixt, O, Taylor, M.E, Weis, W.I, Drickamer, K. | | Deposit date: | 2004-03-05 | | Release date: | 2004-06-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural basis for distinct ligand-binding and targeting properties of the receptors
DC-SIGN and DC-SIGNR
Nat.Struct.Mol.Biol., 11, 2004
|
|
7M7X
 
 | |
5ILF
 
 | | The X-ray structure of the adduct formed in the reaction between hen egg white lysozyme and compound 4, a platin(II) compound containing a O, S bidentate ligand | | Descriptor: | ACETATE ION, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Merlino, A, Ferraro, G. | | Deposit date: | 2016-03-04 | | Release date: | 2016-12-07 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Platinum(ii) O,S complexes as potential metallodrugs against Cisplatin resistance.
Dalton Trans, 45, 2016
|
|
7LVG
 
 | |
5W3Q
 
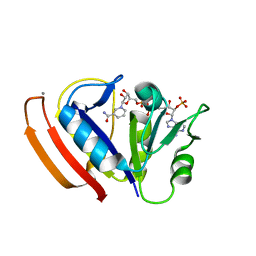 | | L28F E.coli DHFR in complex with NADPH | | Descriptor: | CALCIUM ION, Dihydrofolate reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Oyen, D, Wright, P.E, Wilson, I.A. | | Deposit date: | 2017-06-08 | | Release date: | 2017-08-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.401 Å) | | Cite: | Defining the Structural Basis for Allosteric Product Release from E. coli Dihydrofolate Reductase Using NMR Relaxation Dispersion.
J. Am. Chem. Soc., 139, 2017
|
|
7LOI
 
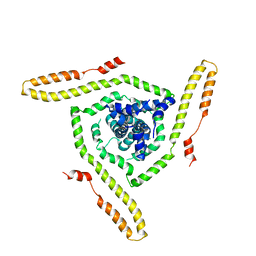 | | Model of the HIV-1 gp41 membrane-proximal external region, transmembrane domain and cytoplasmic tail | | Descriptor: | Transmembrane protein gp41 | | Authors: | Piai, A, Fu, Q, Sharp, A.K, Bighi, B, Brown, A.M, Chou, J.J. | | Deposit date: | 2021-02-10 | | Release date: | 2021-04-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR Model of the Entire Membrane-Interacting Region of the HIV-1 Fusion Protein and Its Perturbation of Membrane Morphology.
J.Am.Chem.Soc., 143, 2021
|
|
5ILN
 
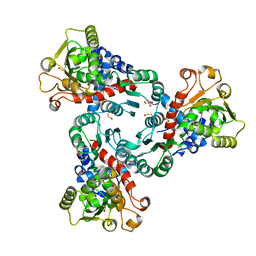 | | Crystal structure of Aspartate Transcarbamoylase from Plasmodium falciparum (PfATC) with bound citrate | | Descriptor: | Aspartate carbamoyltransferase, CITRATE ANION, GLYCEROL, ... | | Authors: | Lunev, S, Bosch, S.S, Batista, F.D.A, Wrenger, C, Groves, M.R. | | Deposit date: | 2016-03-04 | | Release date: | 2017-03-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Crystal structure of truncated Aspartate Transcarbamoylase from Plasmodium falciparum
To be published
|
|
7LXC
 
 | |
5VIP
 
 | |
5VJ6
 
 | |
