1WD3
 
 | | Crystal structure of arabinofuranosidase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, alpha-L-arabinofuranosidase B | | Authors: | Miyanaga, A, Koseki, T, Matsuzawa, H, Wakagi, T, Shoun, H, Fushinobu, S. | | Deposit date: | 2004-05-11 | | Release date: | 2004-09-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of a family 54 alpha-L-arabinofuranosidase reveals a novel carbohydrate-binding module that can bind arabinose
J.Biol.Chem., 279, 2004
|
|
6I7X
 
 | | Crystal Structure of the first bromodomain of BRD4 in complex with RT53 | | Descriptor: | 1,2-ETHANEDIOL, 2-[(4~{S})-6-(4-chlorophenyl)-8-methoxy-1-methyl-4~{H}-[1,2,4]triazolo[4,3-a][1,4]benzodiazepin-4-yl]-1-[4-(dimethylamino)piperidin-1-yl]ethanone, Bromodomain-containing protein 4 | | Authors: | Picaud, S, Traquete, R, Bernardes, G.J.L, Newman, J, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Filippakopoulos, P, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-11-19 | | Release date: | 2019-11-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal Structure of the first bromodomain of BRD4 in complex with RT53
To Be Published
|
|
6I81
 
 | | Crystal Structure of the second bromodomain of BRD2 in complex with RT56 | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Bromodomain-containing protein 2, ... | | Authors: | Picaud, S, Traquete, R, Bernardes, G.J.L, Newman, J, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Filippakopoulos, P, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-11-19 | | Release date: | 2019-11-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Crystal Structure of the second bromodomain of BRD2 in complex with RT56
To Be Published
|
|
5QCV
 
 | | Crystal structure of BACE complex with BMC023 | | Descriptor: | (10S,13S)-13-[(1R)-1-hydroxy-2-({[3-(propan-2-yl)phenyl]methyl}amino)ethyl]-9,10-dimethyl-2-oxa-9,12-diazabicyclo[13.3.1]nonadeca-1(19),15,17-triene-8,11-dione, Beta-secretase 1 | | Authors: | Rondeau, J.M, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-12-01 | | Release date: | 2020-06-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | D3R grand challenge 4: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J.Comput.Aided Mol.Des., 34, 2020
|
|
4A00
 
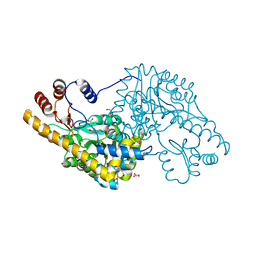 | | Structure of an engineered aspartate aminotransferase | | Descriptor: | ALANYL-PYRIDOXAL-5'-PHOSPHATE, ASPARTATE AMINOTRANSFERASE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Fernandez, F.J, deVries, D, Pena-Soler, E, Coll, M, Christen, P, Gehring, H, Vega, M.C. | | Deposit date: | 2011-09-06 | | Release date: | 2011-12-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Structure and Mechanism of a Cysteine Sulfinate Desulfinase Engineered on the Aspartate Aminotransferase Scaffold.
Biocim.Biophys.Acta, 1824, 2011
|
|
5QDZ
 
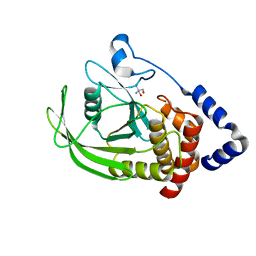 | | PanDDA analysis group deposition -- Crystal structure of PTP1B in complex with compound_FMOPL000435a | | Descriptor: | (azepan-1-yl)(2H-1,3-benzodioxol-5-yl)methanone, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Keedy, D.A, Hill, Z.B, Biel, J.T, Kang, E, Rettenmaier, T.J, Brandao-Neto, J, von Delft, F, Wells, J.A, Fraser, J.S. | | Deposit date: | 2018-08-30 | | Release date: | 2018-10-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.141 Å) | | Cite: | An expanded allosteric network in PTP1B by multitemperature crystallography, fragment screening, and covalent tethering.
Elife, 7, 2018
|
|
1LMQ
 
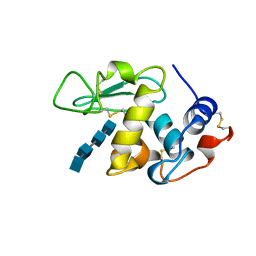 | |
3QR6
 
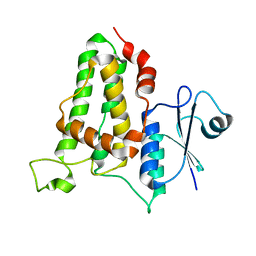 | |
1A0Q
 
 | | 29G11 COMPLEXED WITH PHENYL [1-(1-N-SUCCINYLAMINO)PENTYL] PHOSPHONATE | | Descriptor: | 29G11 FAB (HEAVY CHAIN), 29G11 FAB (LIGHT CHAIN), PHENYL[1-(N-SUCCINYLAMINO)PENTYL]PHOSPHONATE, ... | | Authors: | Buchbinder, J.L, Stephenson, R.C, Scanlan, T.S, Fletterick, R.J. | | Deposit date: | 1997-12-05 | | Release date: | 1999-03-02 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A comparison of the crystallographic structures of two catalytic antibodies with esterase activity.
J.Mol.Biol., 282, 1998
|
|
5QF2
 
 | | PanDDA analysis group deposition -- Crystal structure of PTP1B in complex with compound_FMOMB000187a | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-(5,6-dichloro-1H-benzimidazol-1-yl)butan-1-ol, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Keedy, D.A, Hill, Z.B, Biel, J.T, Kang, E, Rettenmaier, T.J, Brandao-Neto, J, von Delft, F, Wells, J.A, Fraser, J.S. | | Deposit date: | 2018-08-30 | | Release date: | 2018-10-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.772 Å) | | Cite: | An expanded allosteric network in PTP1B by multitemperature crystallography, fragment screening, and covalent tethering.
Elife, 7, 2018
|
|
3QRE
 
 | |
2X95
 
 | | Crystal structure of AnCE-lisinopril-tryptophan analogue, lisW-S complex | | Descriptor: | (S)-1-N2-(1-CARBOXY-3-PHENYLPROPYL)-L-LYSYL-L-TRYPTOPHAN, 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Akif, M, Georgiadis, D, Mahajan, A, Dive, V, Sturrock, E.D, Isaac, R.E, Acharya, K.R. | | Deposit date: | 2010-03-14 | | Release date: | 2010-06-02 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | High Resolution Crystal Structures of Drosophila Melanogaster Angiotensin Converting Enzyme in Complex with Novel Inhibitors and Anti- Hypertensive Drugs.
J.Mol.Biol., 400, 2010
|
|
2F25
 
 | | Crystal Structure of the Human Sialidase Neu2 E111Q Mutant in Complex with DANA Inhibitor | | Descriptor: | 2-DEOXY-2,3-DEHYDRO-N-ACETYL-NEURAMINIC ACID, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Sialidase 2 | | Authors: | Chavas, L.M.G, Kato, R, Fusi, P, Tringali, C, Venerando, B, Tettamanti, G, Monti, E, Wakatsuki, S. | | Deposit date: | 2005-11-15 | | Release date: | 2006-11-21 | | Last modified: | 2025-06-04 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure of the Human Sialidase Neu2 E111Q Mutant in Complex with DANA Inhibitor
To be Published
|
|
3QUP
 
 | |
4D5J
 
 | | Hypocrea jecorina cellobiohydrolase Cel7A E217Q soaked with xylotriose. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CELLULOSE 1,4-BETA-CELLOBIOSIDASE, COBALT (II) ION, ... | | Authors: | Momeni, M.H, Ubhayasekera, W, Stahlberg, J, Hansson, H. | | Deposit date: | 2014-11-05 | | Release date: | 2015-03-25 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Insights Into the Inhibition of Cellobiohydrolase Cel7A by Xylooligosaccharides.
FEBS J., 282, 2015
|
|
3QU2
 
 | | Crystal structure of pyrophosphatase from bacteroides thetaiotaomicron, a closed cap conformation | | Descriptor: | CHLORIDE ION, CITRIC ACID, GLYCEROL, ... | | Authors: | Patskovsky, Y, Huang, H, Toro, R, Gerlt, J.A, Burley, S.K, Dunaway-Mariano, D, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC), Enzyme Function Initiative (EFI) | | Deposit date: | 2011-02-23 | | Release date: | 2011-04-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Divergence of Structure and Function in the Haloacid Dehalogenase Enzyme Superfamily: Bacteroides thetaiotaomicron BT2127 Is an Inorganic Pyrophosphatase.
Biochemistry, 50, 2011
|
|
4QWP
 
 | | co-crystal structure of chitosanase OU01 with substrate | | Descriptor: | 2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose, 2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose, ACETATE ION, ... | | Authors: | Lyu, Q, Liu, W, Han, B. | | Deposit date: | 2014-07-17 | | Release date: | 2015-07-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and biochemical insights into the degradation mechanism of chitosan by chitosanase OU01.
Biochim.Biophys.Acta, 1850, 2015
|
|
2YFR
 
 | | Crystal structure of inulosucrase from Lactobacillus johnsonii NCC533 | | Descriptor: | ACETATE ION, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Pijning, T, Anwar, M.A, Leemhuis, H, Kralj, S, Dijkhuizen, L, Dijkstra, B.W. | | Deposit date: | 2011-04-07 | | Release date: | 2011-08-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structure of Inulosucrase from Lactobacillus: Insights Into the Substrate Specificity and Product Specificity of Gh68 Fructansucrases.
J.Mol.Biol., 412, 2011
|
|
2VQ6
 
 | |
3PC8
 
 | | X-ray crystal structure of the heterodimeric complex of XRCC1 and DNA ligase III-alpha BRCT domains. | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DNA ligase 3, DNA repair protein XRCC1, ... | | Authors: | Cuneo, M.J, Krahn, J.M, London, R.E. | | Deposit date: | 2010-10-21 | | Release date: | 2011-06-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | The structural basis for partitioning of the XRCC1/DNA ligase III-{alpha} BRCT-mediated dimer complexes.
Nucleic Acids Res., 39, 2011
|
|
4BL9
 
 | | Crystal structure of full-length human Suppressor of fused (SUFU) mutant lacking a regulatory subdomain (crystal form I) | | Descriptor: | MALTOSE-BINDING PERIPLASMIC PROTEIN, SUPPRESSOR OF FUSED HOMOLOG, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Cherry, A.L, Finta, C, Karlstrom, M, Toftgard, R, Jovine, L. | | Deposit date: | 2013-05-02 | | Release date: | 2013-11-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Basis of Sufu-GLI Interaction in Hedgehog Signalling Regulation
Acta Crystallogr.,Sect.D, 69, 2013
|
|
1ODZ
 
 | | Expansion of the glycosynthase repertoire to produce defined manno-oligosaccharides | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Mannan endo-1,4-beta-mannosidase, SODIUM ION, ... | | Authors: | Jahn, M, Stoll, D, Warren, R.A.J, Szabo, L, Singh, P, Gilbert, H.J, Ducros, V.M.A, Davies, G.J, Withers, S.G. | | Deposit date: | 2003-03-17 | | Release date: | 2003-07-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Expansion of the Glycosynthase Repertoire to Produce Defined Manno-Oligosaccharides
Chem.Commun.(Camb.), 12, 2003
|
|
1ZRF
 
 | | 4 crystal structures of CAP-DNA with all base-pair substitutions at position 6, CAP-[6C;17G]ICAP38 DNA | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, 5'-D(*AP*TP*TP*TP*CP*GP*AP*AP*AP*AP*AP*TP*GP*CP*GP*AP*T)-3', 5'-D(*CP*TP*AP*GP*AP*TP*CP*GP*CP*AP*TP*TP*TP*TP*TP*CP*GP*AP*AP*AP*T)-3', ... | | Authors: | Berman, H.M, Napoli, A.A. | | Deposit date: | 2005-05-19 | | Release date: | 2006-03-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Indirect readout of DNA sequence at the primary-kink site in the CAP-DNA complex: recognition of pyrimidine-purine and purine-purine steps.
J.Mol.Biol., 357, 2006
|
|
4AUJ
 
 | | FimH lectin domain co-crystal with a alpha-D-mannoside O-linked to para hydroxypropargyl phenyl | | Descriptor: | 4-(3-hydroxyprop-1-yn-1-yl)phenyl alpha-D-mannopyranoside, FIMH | | Authors: | Bouckaert, J, Touaibia, M, Roos, G, Shiao, T.C, Wang, Q, Papadopoulos, A, Roy, R. | | Deposit date: | 2012-05-17 | | Release date: | 2013-05-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.527 Å) | | Cite: | Validation of Reactivity Descriptors to Assess the Aromatic Stacking within the Tyrosine Gate of Fimh.
Acs Med.Chem.Lett., 4, 2013
|
|
4AVK
 
 | | Structure of trigonal FimH lectin domain crystal soaked with an alpha- D-mannoside O-linked to propynyl pyridine at 2.4A resolution | | Descriptor: | 3-(pyridin-3-yl)prop-2-yn-1-yl alpha-D-mannopyranoside, FIMH, NICKEL (II) ION, ... | | Authors: | Wellens, A, Lahmann, M, Touaibia, M, Vaucher, J, Oscarson, S, Roy, R, Remaut, H, Bouckaert, J. | | Deposit date: | 2012-05-26 | | Release date: | 2012-06-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Tyrosine Gate as a Potential Entropic Lever in the Receptor-Binding Site of the Bacterial Adhesin Fimh.
Biochemistry, 51, 2012
|
|
