7RCE
 
 | |
1M8U
 
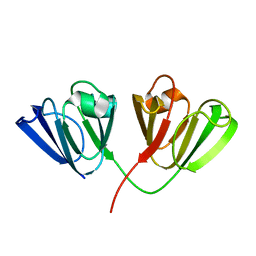 | | Crystal Structure of Bovine gamma-E at 1.65 Ang Resolution | | Descriptor: | gamma-E | | Authors: | Mayer, C, Agueznay, N, Skouri-Panet, F, Prat, K, Putilina, T, Biarrotte-Sorin, S, Tardieu, A. | | Deposit date: | 2002-07-26 | | Release date: | 2003-08-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal Structure of Bovine gamma-E
To be Published
|
|
9G3M
 
 | |
7RCC
 
 | |
9GQU
 
 | |
1M9B
 
 | | Crystal structure of the 26 kDa glutathione S-transferase from Schistosoma japonicum complexed with gamma-glutamyl[S-(2-iodobenzyl)cysteinyl]glycine | | Descriptor: | GAMMA-GLUTAMYL[S-(2-IODOBENZYL)CYSTEINYL]GLYCINE, Glutathione S-Transferase 26 kDa | | Authors: | Cardoso, R.M.F, Daniels, D.S, Bruns, C.M, Tainer, J.A. | | Deposit date: | 2002-07-28 | | Release date: | 2003-03-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Characterization of the electrophile
binding site and substrate binding
mode of the 26-kDa glutathione
S-transferase from Schistosoma
japonicum
PROTEINS: STRUCT.,FUNCT.,GENET., 51, 2003
|
|
1M9G
 
 | | Solution structure of G16A-MNEI, a structural mutant of single chain monellin MNEI | | Descriptor: | Monellin chain B and Monellin chain A | | Authors: | Spadaccini, R, Trabucco, F, Saviano, G, Picone, D, Crescenzi, O, Tancredi, T, Temussi, P.A. | | Deposit date: | 2002-07-29 | | Release date: | 2003-06-10 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The Mechanism of Interaction of Sweet Proteins with the T1R2-T1R3 Receptor: Evidence from the Solution Structure of G16A-MNEI
J.MOL.BIOL., 328, 2003
|
|
9G3J
 
 | |
1MA9
 
 | | Crystal structure of the complex of human vitamin D binding protein and rabbit muscle actin | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin, Alpha Skeletal Muscle, ... | | Authors: | Verboven, C, Bogaerts, I, Waelkens, E, Rabijns, A, Van Baelen, H, Bouillon, R, De Ranter, C. | | Deposit date: | 2002-08-02 | | Release date: | 2003-02-04 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Actin-DBP: the perfect structural fit?
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1MAV
 
 | | CRYSTAL STRUCTURE OF THE RESPONSE REGULATOR DIVK AT PH 6.0 IN COMPLEX WITH MN2+ | | Descriptor: | MANGANESE (II) ION, cell division response regulator DivK | | Authors: | Guillet, V, Ohta, N, Cabantous, S, Newton, A, Samama, J.-P, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2002-08-02 | | Release date: | 2002-12-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystallographic and Biochemical Studies of DivK Reveal Novel Features of
an Essential Response Regulator in Caulobacter crescentus.
J.Biol.Chem., 277, 2002
|
|
9GSJ
 
 | | BmrA E504A in complex with Hoechst33342 | | Descriptor: | 2'-(4-ETHOXYPHENYL)-5-(4-METHYL-1-PIPERAZINYL)-2,5'-BI-BENZIMIDAZOLE, Multidrug resistance ABC transporter ATP-binding/permease protein BmrA | | Authors: | Moissonnier, L, Zarkadas, E, Schoehn, G, Falson, P, Chaptal, V. | | Deposit date: | 2024-09-16 | | Release date: | 2025-03-05 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Rhodamine6G and Hœchst33342 narrow BmrA conformational spectrum for a more efficient use of ATP.
Nat Commun, 16, 2025
|
|
9G3N
 
 | |
1MD4
 
 | | A folding mutant of human class pi glutathione transferase, created by mutating glycine 146 of the wild-type protein to valine | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLUTATHIONE, pi glutathione transferase | | Authors: | Kong, G.K.-W, Dragani, B, Aceto, A, Cocco, R, Mannervik, B, Stenberg, G, McKinstry, W.J, Polekhina, G, Parker, M.W. | | Deposit date: | 2002-08-06 | | Release date: | 2002-08-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Contribution of Glycine 146 to a Conserved Folding Module Affecting Stability and Refolding of Human Glutathione Transferase P1-1
J.Biol.Chem., 278, 2003
|
|
1MDO
 
 | | Crystal structure of ArnB aminotransferase with pyridomine 5' phosphate | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, ArnB aminotransferase | | Authors: | Noland, B.W, Newman, J.M, Hendle, J, Badger, J, Christopher, J.A, Tresser, J, Buchanan, M.D, Wright, T, Rutter, M.E, Sanderson, W.E, Muller-Dieckmann, H.-J, Gajiwala, K, Sauder, J.M, Buchanan, S.G. | | Deposit date: | 2002-08-07 | | Release date: | 2002-12-11 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural studies of Salmonella typhimurium ArnB (PmrH) aminotransferase: A 4-amino-4-deoxy-L-arabinose lipopolysaccharide modifying enzyme
Structure, 10, 2002
|
|
9G8I
 
 | | Sumo-Darpin-A10-complex | | Descriptor: | DARPin, Ubiquitin-like protein SMT3 | | Authors: | Cakilkaya, B, Wolf, E, Boergel, A. | | Deposit date: | 2024-07-23 | | Release date: | 2025-03-05 | | Last modified: | 2025-06-11 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Custom affinity probes reveal DNA-damage-induced, ssDNA-independent chromatin SUMOylation in budding yeast.
Cell Rep, 44, 2025
|
|
1ME3
 
 | |
9GJT
 
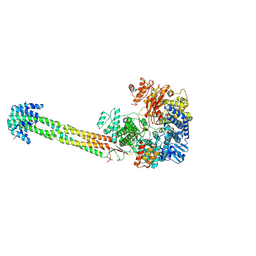 | | Structure of Nipah Virus RNA Polymerase Complex - Apo state | | Descriptor: | Phosphoprotein, RNA-directed RNA polymerase L, ZINC ION | | Authors: | Sala, F, Ditter, K, Dybkov, O, Urlaub, H, Hillen, H.S. | | Deposit date: | 2024-08-22 | | Release date: | 2025-03-05 | | Last modified: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural basis of Nipah virus RNA synthesis.
Nat Commun, 16, 2025
|
|
7RVG
 
 | |
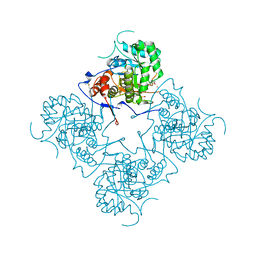
1MEH
 
 | |
9GMB
 
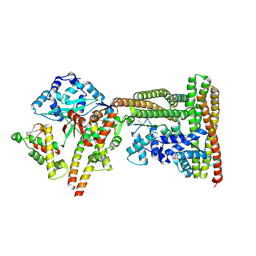 | | MukEF in complex with the phage protein gp5.9 | | Descriptor: | Chromosome partition protein MukE, Chromosome partition protein MukF, Probable RecBCD inhibitor gp5.9 | | Authors: | Burmann, F, Wilkinson, O, Kimanius, D, Dillingham, M, Lowe, J. | | Deposit date: | 2024-08-28 | | Release date: | 2025-03-26 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Mechanism of DNA capture by the MukBEF SMC complex and its inhibition by a viral DNA mimic.
Cell, 188, 2025
|
|
1MET
 
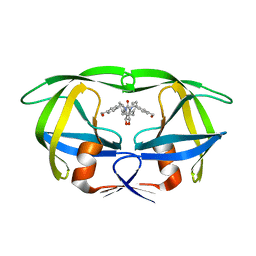 | | HIV-1 MUTANT (V82F) PROTEASE COMPLEXED WITH DMP323 | | Descriptor: | HIV-1 PROTEASE, [4-R-(-4-ALPHA,5-ALPHA,6-BETA,7-BETA)]-HEXAHYDRO-5,6-BIS(HYDROXY)-[1,3-BIS([4-HYDROXYMETHYL-PHENYL]METHYL)-4,7-BIS(PHEN YLMETHYL)]-2H-1,3-DIAZEPINONE | | Authors: | Ala, P, Chang, C.-H. | | Deposit date: | 1997-04-11 | | Release date: | 1998-04-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular basis of HIV-1 protease drug resistance: structural analysis of mutant proteases complexed with cyclic urea inhibitors.
Biochemistry, 36, 1997
|
|
7R6G
 
 | | Crystal structure of DfrA5 dihydrofolate reductase in complex with TRIMETHOPRIM and NADPH | | Descriptor: | Dihydrofolate reductase type 5, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, SULFATE ION, ... | | Authors: | Estrada, A, Wright, D, Krucinska, J, Erlandsen, H. | | Deposit date: | 2021-06-22 | | Release date: | 2022-06-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Structure-guided functional studies of plasmid-encoded dihydrofolate reductases reveal a common mechanism of trimethoprim resistance in Gram-negative pathogens.
Commun Biol, 5, 2022
|
|
9GB2
 
 | | Extended phiCD508 baseplate | | Descriptor: | gp55 - Tail sheath protein, gp56 - Tail tube protein, gp61 - Tail tube initiator, ... | | Authors: | Wilson, J.S, Fagan, R.P, Bullough, P.A. | | Deposit date: | 2024-07-29 | | Release date: | 2025-03-26 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (3.43 Å) | | Cite: | Molecular mechanism of bacteriophage contraction structure of an S-layer-penetrating bacteriophage.
Life Sci Alliance, 8, 2025
|
|
1MF6
 
 | |
9GIO
 
 | |
