2ZKF
 
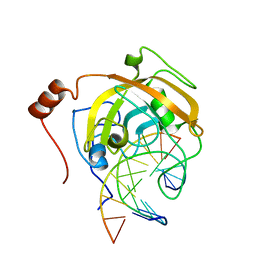 | | Crystal structure of the SRA domain of mouse Np95 in complex with hemi-methylated CpG DNA | | Descriptor: | DNA (5'-D(*DCP*DTP*DAP*DTP*DCP*(5CM)P*DGP*DGP*DTP*DGP*DA)-3'), DNA (5'-D(P*DCP*DAP*DCP*DCP*DGP*DGP*DAP*DTP*DAP*DGP*DA)-3'), E3 ubiquitin-protein ligase UHRF1 | | Authors: | Arita, K, Ariyoshi, M, Tochio, H, Nakamura, Y, Shirakawa, M. | | Deposit date: | 2008-03-19 | | Release date: | 2008-09-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Recognition of hemi-methylated DNA by the SRA protein UHRF1 by a base-flipping mechanism
Nature, 455, 2008
|
|
5WFY
 
 | |
3W1O
 
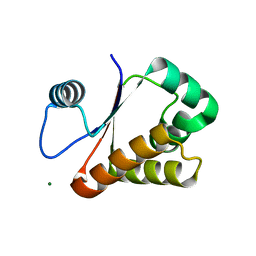 | | Neisseria DNA mimic protein DMP12 | | Descriptor: | DNA mimic protein DMP12, MAGNESIUM ION | | Authors: | Wang, H.C, Ko, T.P, Wu, M.L, Wang, A.H.J. | | Deposit date: | 2012-11-19 | | Release date: | 2013-04-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Neisseria conserved hypothetical protein DMP12 is a DNA mimic that binds to histone-like HU protein
Nucleic Acids Res., 41, 2013
|
|
3RHI
 
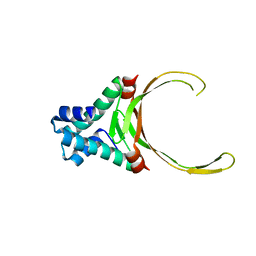 | | DNA-binding protein HU from Bacillus anthracis | | Descriptor: | DNA-binding protein HU | | Authors: | Osipiuk, J, Makowska-Grzyska, M, Hasseman, J, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-04-11 | | Release date: | 2011-04-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | DNA-binding protein HU from Bacillus anthracis.
To be Published
|
|
5NWA
 
 | |
5NW9
 
 | |
1BNP
 
 | | NMR SOLUTION STRUCTURE OF THE N-TERMINAL DOMAIN OF DNA POLYMERASE BETA, 55 STRUCTURES | | Descriptor: | DNA POLYMERASE BETA | | Authors: | Liu, D.-J, Prasad, R, Wilson, S.H, Derose, E.F, Mullen, G.P. | | Deposit date: | 1996-04-25 | | Release date: | 1996-12-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional solution structure of the N-terminal domain of DNA polymerase beta and mapping of the ssDNA interaction interface.
Biochemistry, 35, 1996
|
|
1BNO
 
 | | NMR SOLUTION STRUCTURE OF THE N-TERMINAL DOMAIN OF DNA POLYMERASE BETA, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | DNA POLYMERASE BETA | | Authors: | Liu, D.-J, Prasad, R, Wilson, S.H, Derose, E.F, Mullen, G.P. | | Deposit date: | 1996-04-25 | | Release date: | 1996-12-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional solution structure of the N-terminal domain of DNA polymerase beta and mapping of the ssDNA interaction interface.
Biochemistry, 35, 1996
|
|
1JN3
 
 | |
6FWS
 
 | | Structure of DinG in complex with ssDNA and ADPBeF | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent DNA helicase DinG, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Cheng, K, Wigley, D. | | Deposit date: | 2018-03-07 | | Release date: | 2018-12-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | DNA translocation mechanism of an XPD family helicase.
Elife, 7, 2018
|
|
4M9V
 
 | | Zfp57 mutant (E182Q) in complex with 5-carboxylcytosine DNA | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Liu, Y, Olanrewaju, Y.O, Zhang, X, Cheng, X. | | Deposit date: | 2013-08-15 | | Release date: | 2013-11-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (0.969 Å) | | Cite: | DNA recognition of 5-carboxylcytosine by a zfp57 mutant at an atomic resolution of 0.97 angstrom.
Biochemistry, 52, 2013
|
|
6FWR
 
 | | Structure of DinG in complex with ssDNA | | Descriptor: | ATP-dependent DNA helicase DinG, DNA (5'-D(*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*T)-3'), IRON/SULFUR CLUSTER | | Authors: | Cheng, K, Wigley, D.B. | | Deposit date: | 2018-03-07 | | Release date: | 2018-12-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | DNA translocation mechanism of an XPD family helicase.
Elife, 7, 2018
|
|
6MII
 
 | | Crystal structure of minichromosome maintenance protein MCM/DNA complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (5'-D(P*TP*TP*TP*TP*TP*TP*TP*TP*TP*T)-3'), MAGNESIUM ION, ... | | Authors: | Enemark, E.J, Meagher, M, Epling, L.B. | | Deposit date: | 2018-09-19 | | Release date: | 2019-07-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | DNA translocation mechanism of the MCM complex and implications for replication initiation.
Nat Commun, 10, 2019
|
|
8XIC
 
 | |
5A9J
 
 | | Crystal structure of the Helicase domain of human DNA polymerase theta, apo-form | | Descriptor: | DNA POLYMERASE THETA | | Authors: | Newman, J.A, Cooper, C.D.O, Aitkenhead, H, Pinkas, D.M, Kupinska, K, Burgess-Brown, N, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2015-07-21 | | Release date: | 2015-12-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.55 Å) | | Cite: | Structure of the Helicase Domain of DNA Polymerase Theta Reveals a Possible Role in the Microhomology-Mediated End-Joining Pathway.
Structure, 23, 2015
|
|
8VPI
 
 | |
8VPG
 
 | |
8VPH
 
 | |
8Z79
 
 | |
5JGH
 
 | | Crystal structure of the mitochondrial DNA packaging protein Abf2p in complex with DNA at 2.6 Angstrom resolution | | Descriptor: | ACETATE ION, ARS-binding factor 2, mitochondrial, ... | | Authors: | Chakraborty, A, Lyonnais, S, Sola, M. | | Deposit date: | 2016-04-20 | | Release date: | 2017-02-08 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | DNA structure directs positioning of the mitochondrial genome packaging protein Abf2p.
Nucleic Acids Res., 45, 2017
|
|
8Y1I
 
 | |
5JH0
 
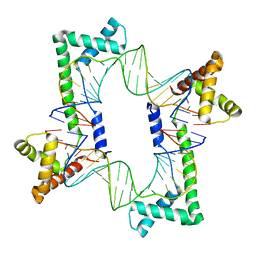 | | Crystal structure of the mitochondrial DNA packaging protein Abf2p in complex with DNA at 2.18 Angstrom resolution | | Descriptor: | ARS-binding factor 2, mitochondrial, DNA (5'-D(*AP*AP*TP*AP*AP*TP*AP*AP*AP*TP*TP*AP*TP*AP*TP*AP*AP*TP*AP*TP*AP*A)-3'), ... | | Authors: | Chakraborty, A, Lyonnais, S, Sola, M. | | Deposit date: | 2016-04-20 | | Release date: | 2017-02-08 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | DNA structure directs positioning of the mitochondrial genome packaging protein Abf2p.
Nucleic Acids Res., 45, 2017
|
|
8VDS
 
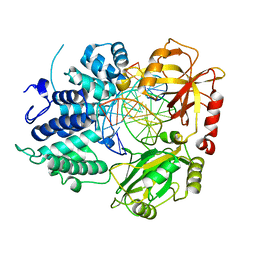 | | DNA Ligase 1 with nick DNA 3'rG:C | | Descriptor: | DNA (5'-D(*GP*TP*CP*CP*GP*AP*CP*CP*AP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), DNA ligase 1, DNA/RNA (5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*T)-R(P*G)-D(P*GP*TP*CP*GP*GP*AP*C)-3') | | Authors: | KanalElamparithi, B, Gulkis, M, Caglayan, M. | | Deposit date: | 2023-12-17 | | Release date: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Structures of LIG1 provide a mechanistic basis for understanding a lack of sugar discrimination against a ribonucleotide at the 3'-end of nick DNA.
J.Biol.Chem., 300, 2024
|
|
8VDT
 
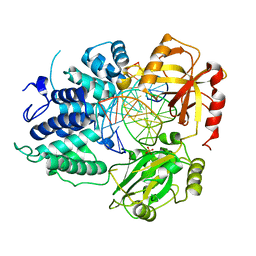 | | DNA Ligase 1 with nick DNA 3'rA:T | | Descriptor: | DNA (5'-D(*GP*TP*CP*CP*GP*AP*CP*TP*AP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), DNA ligase 1, DNA/RNA (5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*T)-R(P*A)-D(P*GP*TP*CP*GP*GP*AP*C)-3'), ... | | Authors: | KanalElamparithi, B, Gulkis, M, Caglayan, M. | | Deposit date: | 2023-12-17 | | Release date: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Structures of LIG1 provide a mechanistic basis for understanding a lack of sugar discrimination against a ribonucleotide at the 3'-end of nick DNA.
J.Biol.Chem., 300, 2024
|
|
4LYL
 
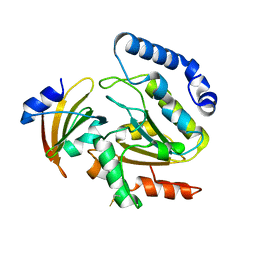 | | Crystal structure of uracil-DNA glycosylase from cod (Gadus morhua) in complex with the proteinaceous inhibitor UGI | | Descriptor: | Uracil-DNA glycosylase, Uracil-DNA glycosylase inhibitor | | Authors: | Assefa, N.G, Niiranen, L.M.K, Johnson, K.A, Leiros, H.-K.S, Smalas, A.O, Willassen, N.P, Moe, E. | | Deposit date: | 2013-07-31 | | Release date: | 2014-08-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structural and biophysical analysis of interactions between cod and human uracil-DNA N-glycosylase (UNG) and UNG inhibitor (Ugi).
Acta Crystallogr.,Sect.D, 70, 2014
|
|
