2EPL
 
 | | N-acetyl-B-D-glucosaminidase (GCNA) from Streptococcus gordonii | | Descriptor: | GLYCEROL, N-acetyl-beta-D-glucosaminidase, SULFATE ION | | Authors: | Langley, D.B, Harty, D.W.S, Guss, J.M, Collyer, C.A. | | Deposit date: | 2007-03-30 | | Release date: | 2008-03-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure of N-acetyl-beta-D-glucosaminidase (GcnA) from the Endocarditis Pathogen Streptococcus gordonii and its Complex with the Mechanism-based Inhibitor NAG-thiazoline
J.Mol.Biol., 377, 2008
|
|
2EPM
 
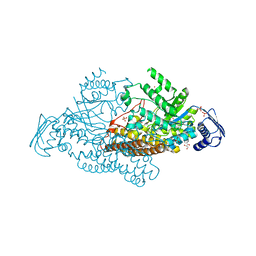 | | N-acetyl-B-D-glucoasminidase (GCNA) from Stretococcus gordonii | | Descriptor: | GLYCEROL, MERCURY (II) ION, N-acetyl-beta-D-glucosaminidase, ... | | Authors: | Langley, D.B, Harty, D.W.S, Guss, J.M, Collyer, C.A. | | Deposit date: | 2007-03-30 | | Release date: | 2008-03-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structure of N-acetyl-beta-D-glucosaminidase (GcnA) from the Endocarditis Pathogen Streptococcus gordonii and its Complex with the Mechanism-based Inhibitor NAG-thiazoline
J.Mol.Biol., 377, 2008
|
|
6JCE
 
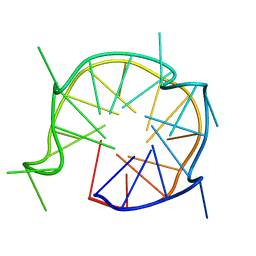 | | NMR solution and X-ray crystal structures of a DNA containing both right-and left-handed parallel-stranded G-quadruplexes | | Descriptor: | 29-mer DNA | | Authors: | Winnerdy, F.R, Bakalar, B, Maity, A, Vandana, J.J, Mechulam, Y, Schmitt, E, Phan, A.T. | | Deposit date: | 2019-01-28 | | Release date: | 2019-07-10 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR solution and X-ray crystal structures of a DNA molecule containing both right- and left-handed parallel-stranded G-quadruplexes.
Nucleic Acids Res., 47, 2019
|
|
2EPK
 
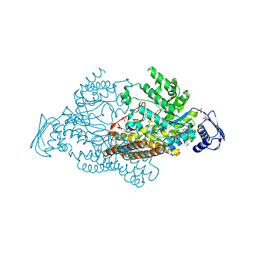 | | N-acetyl-B-D-glucosaminidase (GCNA) from Streptococcus gordonii | | Descriptor: | N-acetyl-beta-D-glucosaminidase, SULFATE ION | | Authors: | Langley, D.B. | | Deposit date: | 2007-03-30 | | Release date: | 2008-03-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of N-acetyl-beta-D-glucosaminidase (GcnA) from the Endocarditis Pathogen Streptococcus gordonii and its Complex with the Mechanism-based Inhibitor NAG-thiazoline
J.Mol.Biol., 377, 2008
|
|
3RAZ
 
 | |
3TQ5
 
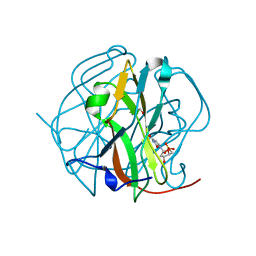 | |
3U97
 
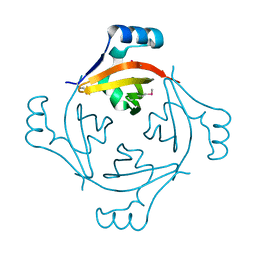 | |
5OV6
 
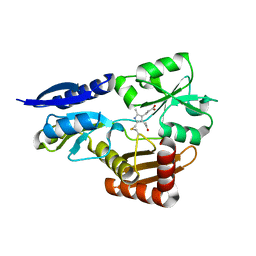 | | Bacillus megaterium porphobilinogen deaminase D82N mutant | | Descriptor: | 3-[4-(2-hydroxy-2-oxoethyl)-2,5-dimethyl-1~{H}-pyrrol-3-yl]propanoic acid, Porphobilinogen deaminase | | Authors: | Guo, J, Erskine, P, Coker, A.R, Wood, S.P, Cooper, J.B. | | Deposit date: | 2017-08-28 | | Release date: | 2017-09-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structural studies of domain movement in active-site mutants of porphobilinogen deaminase from Bacillus megaterium.
Acta Crystallogr F Struct Biol Commun, 73, 2017
|
|
2HIU
 
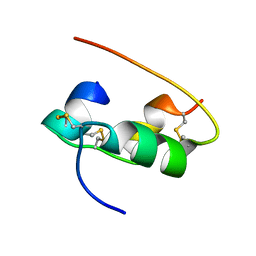 | | NMR STRUCTURE OF HUMAN INSULIN IN 20% ACETIC ACID, ZINC-FREE, 10 STRUCTURES | | Descriptor: | INSULIN | | Authors: | Hua, Q.X, Gozani, S.N, Chance, R.E, Hoffmann, J.A, Frank, B.H, Weiss, M.A. | | Deposit date: | 1996-10-08 | | Release date: | 1997-04-01 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Structure of a protein in a kinetic trap.
Nat.Struct.Biol., 2, 1995
|
|
3TVY
 
 | |
4IIL
 
 | | Crystal Structure of RfuA (TP0298) of T. pallidum Bound to Riboflavin | | Descriptor: | 1,2-ETHANEDIOL, Membrane lipoprotein TpN38(b), POTASSIUM ION, ... | | Authors: | Brautigam, C.A, Deka, R.K, Norgard, M.V. | | Deposit date: | 2012-12-20 | | Release date: | 2013-02-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Evidence for an ABC-type riboflavin transporter system in pathogenic spirochetes.
MBio, 4, 2013
|
|
2FF7
 
 | | The ABC-ATPase of the ABC-transporter HlyB in the ADP bound state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Alpha-hemolysin translocation ATP-binding protein hlyB | | Authors: | Zaitseva, J, Oswald, C, Jumpertz, T, Jenewein, S, Holland, I.B, Schmitt, L. | | Deposit date: | 2005-12-19 | | Release date: | 2006-08-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A structural analysis of asymmetry required for catalytic activity of an ABC-ATPase domain dimer.
Embo J., 25, 2006
|
|
1AGL
 
 | | STRUCTURE OF A DNA-BISDAUNOMYCIN COMPLEX | | Descriptor: | 4-METHYLBENZYL-N-BIS[DAUNOMYCIN], DNA (5'-D(*CP*GP*AP*TP*CP*G)-3') | | Authors: | Hu, G.G, Shui, X, Leng, F, Priebe, W, Chaires, J.B, Williams, L.D. | | Deposit date: | 1997-03-25 | | Release date: | 1997-04-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of a DNA-bisdaunomycin complex.
Biochemistry, 36, 1997
|
|
2FEI
 
 | | Solution structure of the second SH3 domain of Human CMS protein | | Descriptor: | CD2-associated protein | | Authors: | Yao, B, Dai, H, Jiao, Y, Wu, J, Shi, Y. | | Deposit date: | 2005-12-15 | | Release date: | 2006-12-05 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the second SH3 domain of human CMS and a newly identified binding site at the C-terminus of c-Cbl
Biochim.Biophys.Acta, 1774, 2007
|
|
1VEE
 
 | | NMR structure of the hypothetical rhodanese domain At4g01050 from Arabidopsis thaliana | | Descriptor: | proline-rich protein family | | Authors: | Pantoja-Uceda, D, Lopez-Mendez, B, Koshiba, S, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Tanaka, A, Seki, M, Shinozaki, K, Yokoyama, S, Guntert, P, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-03-30 | | Release date: | 2005-01-25 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the rhodanese homology domain At4g01050(175-295) from Arabidopsis thaliana
Protein Sci., 14, 2005
|
|
3TMZ
 
 | | Crystal Structure of P450 2B4(H226Y) in complex with Amlodipine | | Descriptor: | 5-CYCLOHEXYL-1-PENTYL-BETA-D-MALTOSIDE, Amlodipine, Cytochrome P450 2B4, ... | | Authors: | Shah, M.B, Pascual, J, Stout, C.D, Halpert, J.R. | | Deposit date: | 2011-09-01 | | Release date: | 2012-09-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.248 Å) | | Cite: | Conformational Adaptation of Human Cytochrome P450 2B6 and Rabbit Cytochrome P450 2B4 Revealed upon Binding Multiple Amlodipine Molecules.
Biochemistry, 51, 2012
|
|
3RI7
 
 | | Toluene 4 monooxygenase HD Mutant G103L | | Descriptor: | ACETATE ION, FE (III) ION, Toluene-4-monooxygenase system protein A, ... | | Authors: | Bailey, L.J, Elsen, N.L, Fox, B.G. | | Deposit date: | 2011-04-13 | | Release date: | 2012-02-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystallographic analysis of active site contributions to regiospecificity in the diiron enzyme toluene 4-monooxygenase.
Biochemistry, 51, 2012
|
|
2CBN
 
 | |
3TGE
 
 | | A novel series of potent and selective PDE5 inhibitor1 | | Descriptor: | 7-(6-methoxypyridin-3-yl)-3-{[2-(morpholin-4-yl)ethyl]amino}-1-(2-propoxyethyl)pyrido[3,4-b]pyrazin-2(1H)-one, MAGNESIUM ION, ZINC ION, ... | | Authors: | Han, S. | | Deposit date: | 2011-08-17 | | Release date: | 2011-11-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Investigation of the pyrazinones as PDE5 inhibitors: Evaluation of regioisomeric projections into the solvent region.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
2FCI
 
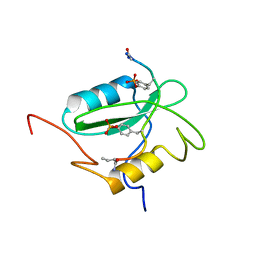 | | Structural basis for the requirement of two phosphotyrosines in signaling mediated by Syk tyrosine kinase | | Descriptor: | C-termainl SH2 domain from phospholipase C-gamma-1 comprising residues 663-759, Doubly phosphorylated peptide derived from Syk kinase comprising residues 338-350 | | Authors: | Groesch, T.D, Zhou, F, Mattila, S, Geahlen, R.L, Post, C.B. | | Deposit date: | 2005-12-12 | | Release date: | 2006-01-31 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the requirement of two phosphotyrosine residues in signaling mediated by syk tyrosine kinase
J.Mol.Biol., 356, 2006
|
|
4JOA
 
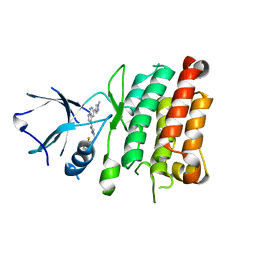 | | Crystal Structure of Human Anaplastic Lymphoma Kinase in complex with 7-azaindole based inhibitor | | Descriptor: | 3-[1-(2,5-difluorobenzyl)-1H-pyrazol-4-yl]-5-(1-methyl-1H-pyrazol-4-yl)-1H-pyrrolo[2,3-b]pyridine, ALK tyrosine kinase receptor | | Authors: | Hosahalli, S, Krishnamurthy, N.R, Lakshminarasimhan, A. | | Deposit date: | 2013-03-18 | | Release date: | 2013-07-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Discovery of 7-azaindole based anaplastic lymphoma kinase (ALK) inhibitors: wild type and mutant (L1196M) active compounds with unique binding mode
Bioorg.Med.Chem.Lett., 23, 2013
|
|
3TQ3
 
 | |
2DMC
 
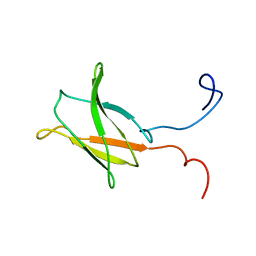 | | Solution structure of the 18th Filamin domain from human Filamin-B | | Descriptor: | Filamin-B | | Authors: | Tomizawa, T, Koshiba, S, Watanabe, S, Harada, T, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-04-20 | | Release date: | 2006-10-20 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the 18th Filamin domain from human Filamin-B
To be Published
|
|
1VDY
 
 | | NMR Structure of the hypothetical ENTH-VHS domain At3g16270 from Arabidopsis thaliana | | Descriptor: | hypothetical protein (RAFL09-17-B18) | | Authors: | Lopez-Mendez, B, Pantoja-Uceda, D, Tomizawa, T, Koshiba, S, Kigawa, T, Shirouzu, M, Terada, T, Inoue, M, Yabuki, T, Aoki, M, Seki, E, Matsuda, T, Hirota, H, Yoshida, M, Tanaka, A, Osanai, T, Seki, M, Shinozaki, K, Yokoyama, S, Guntert, P, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-03-25 | | Release date: | 2005-05-03 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the hypothetical ENTH-VHS domain AT3G16270 from arabidopsis thaliana
To be Published
|
|
3TWZ
 
 | | Phosphorylated Bacillus cereus phosphopentomutase in space group P212121 | | Descriptor: | MANGANESE (II) ION, Phosphopentomutase | | Authors: | Panosian, T.P, Nanneman, D.P, Bachmann, B.O, Iverson, T.M. | | Deposit date: | 2011-09-22 | | Release date: | 2012-02-29 | | Last modified: | 2012-03-21 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Molecular Differences between a Mutase and a Phosphatase: Investigations of the Activation Step in Bacillus cereus Phosphopentomutase.
Biochemistry, 51, 2012
|
|
