2JQ1
 
 | | Phylloseptin-3 | | Descriptor: | Phylloseptin-3 | | Authors: | Resende, J.M, Mendonca Moraes, C, Almeida, F.C.L, Prates, M.V, Cesar, A, Valente, A, Bemquerer, M.P, Pilo-Veloso, D, Bechinger, B. | | Deposit date: | 2007-05-25 | | Release date: | 2008-06-03 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structures of the antimicrobial peptides phylloseptin-1, -2, and -3 and biological activity: The role of charges and hydrogen bonding interactions in stabilizing helix conformations
Peptides, 29, 2008
|
|
3TA8
 
 | | Crystal structure HP-NAP from strain YS39 iron loaded (cocrystallization 5mM) | | Descriptor: | 1,2-ETHANEDIOL, FE (III) ION, Neutrophil-activating protein | | Authors: | Tsuruta, O, Yokoyama, H, Fujii, S. | | Deposit date: | 2011-08-03 | | Release date: | 2012-02-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A new crystal lattice structure of Helicobacter pylori neutrophil-activating protein (HP-NAP)
Acta Crystallogr.,Sect.F, 68, 2012
|
|
2Z4G
 
 | | Histidinol Phosphate Phosphatase from Thermus thermophilus HB8 | | Descriptor: | FE (III) ION, GLYCEROL, Histidinol phosphatase, ... | | Authors: | Omi, R. | | Deposit date: | 2007-06-17 | | Release date: | 2007-11-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Monofunctional Histidinol Phosphate Phosphatase from Thermus thermophilus HB8.
Biochemistry, 46, 2007
|
|
5PQW
 
 | | PanDDA analysis group deposition -- Crystal Structure of BRD1 after initial refinement with no ligand modelled (structure 69) | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 1, SODIUM ION | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-07 | | Release date: | 2017-03-29 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
2JPY
 
 | | Phylloseptin-2 | | Descriptor: | Phylloseptin-2 protein | | Authors: | Resende, J.M, Mendonca Moraes, C, Almeida, F.C.L, Prates, M.V, Cesar, A, Valente, A, Bemquerer, M.P, Pilo-Veloso, D, Bechinger, B. | | Deposit date: | 2007-05-24 | | Release date: | 2008-06-03 | | Last modified: | 2024-10-09 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structures of the antimicrobial peptides phylloseptin-1, -2, and -3 and biological activity: The role of charges and hydrogen bonding interactions in stabilizing helix conformations
Peptides, 29, 2008
|
|
5PRF
 
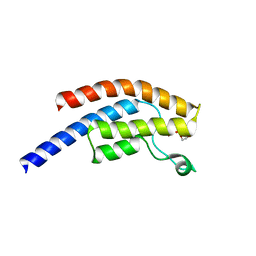 | | PanDDA analysis group deposition -- Crystal Structure of BRD1 after initial refinement with no ligand modelled (structure 87) | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 1, SODIUM ION | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-07 | | Release date: | 2017-03-29 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
5PRU
 
 | | PanDDA analysis group deposition -- Crystal Structure of BRD1 after initial refinement with no ligand modelled (structure 101) | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 1, SODIUM ION | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-07 | | Release date: | 2017-03-29 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
4QVQ
 
 | | yCP beta5-M45I mutant in complex with bortezomib | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, N-[(1R)-1-(DIHYDROXYBORYL)-3-METHYLBUTYL]-N-(PYRAZIN-2-YLCARBONYL)-L-PHENYLALANINAMIDE, ... | | Authors: | Huber, E.M, Heinemeyer, W, Groll, M. | | Deposit date: | 2014-07-15 | | Release date: | 2015-02-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Bortezomib-Resistant Mutant Proteasomes: Structural and Biochemical Evaluation with Carfilzomib and ONX 0914.
Structure, 23, 2015
|
|
4FTT
 
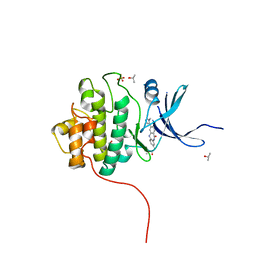 | | Crystal Structure of the CHK1 | | Descriptor: | ISOPROPYL ALCOHOL, SULFATE ION, Serine/threonine-protein kinase Chk1, ... | | Authors: | Kang, Y.N, Stuckey, J.A, Chang, P, Russell, A.J. | | Deposit date: | 2012-06-28 | | Release date: | 2012-08-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of the CHK1
To be Published
|
|
4K0Z
 
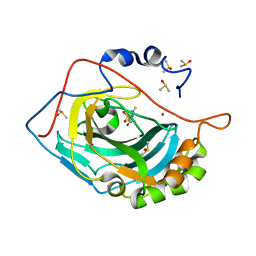 | |
5PS5
 
 | | PanDDA analysis group deposition -- Crystal Structure of BRD1 after initial refinement with no ligand modelled (structure 113) | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 1, SODIUM ION | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-07 | | Release date: | 2017-03-29 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
5PSI
 
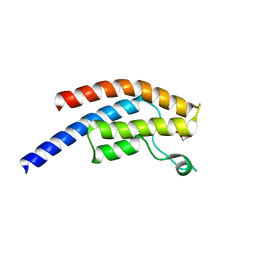 | | PanDDA analysis group deposition -- Crystal Structure of BRD1 after initial refinement with no ligand modelled (structure 126) | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 1, SODIUM ION | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-07 | | Release date: | 2017-03-29 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
5PT0
 
 | | PanDDA analysis group deposition -- Crystal Structure of BRD1 after initial refinement with no ligand modelled (structure 144) | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 1, SODIUM ION | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-07 | | Release date: | 2017-03-29 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
2AW1
 
 | | Carbonic anhydrase inhibitors: Valdecoxib binds to a different active site region of the human isoform II as compared to the structurally related cyclooxygenase II "selective" inhibitor Celecoxib | | Descriptor: | 4-(5-METHYL-3-PHENYLISOXAZOL-4-YL)BENZENESULFONAMIDE, 4-(HYDROXYMERCURY)BENZOIC ACID, Carbonic anhydrase II, ... | | Authors: | Di Fiore, A, Pedone, C, D'Ambrosio, K, Scozzafava, A, De Simone, G, Supuran, C.T. | | Deposit date: | 2005-08-31 | | Release date: | 2006-07-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Carbonic anhydrase inhibitors: Valdecoxib binds to a different active site region of the human isoform II as compared to the structurally related cyclooxygenase II
Bioorg.Med.Chem.Lett., 16, 2006
|
|
5PTG
 
 | | PanDDA analysis group deposition -- Crystal Structure of BRD1 after initial refinement with no ligand modelled (structure 159) | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 1, SODIUM ION | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-07 | | Release date: | 2017-03-29 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
5PTZ
 
 | | PanDDA analysis group deposition -- Crystal Structure of BRD1 after initial refinement with no ligand modelled (structure 177) | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 1, SODIUM ION | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-07 | | Release date: | 2017-03-29 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
2ZAE
 
 | | Crystal structure of protein Ph1601p in complex with protein Ph1771p of archaeal ribonuclease P from Pyrococcus horikoshii OT3 | | Descriptor: | GLYCEROL, NITRATE ION, Ribonuclease P protein component 1, ... | | Authors: | Honda, T, Kakuta, Y, Kimura, M. | | Deposit date: | 2007-10-04 | | Release date: | 2008-10-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structure of an archaeal homolog of the human protein complex Rpp21-Rpp29 that is a key core component for the assembly of active ribonuclease P.
J.Mol.Biol., 384, 2008
|
|
4B1F
 
 | | Design of Inhibitors of Helicobacter pylori Glutamate Racemase as Selective Antibacterial Agents: Incorporation of Imidazoles onto a Core Pyrazolopyrimidinedione Scaffold to Improve Bioavailabilty | | Descriptor: | 5-methyl-3-(1-methyl-1H-imidazol-5-yl)-7-(2-methylpropyl)-2-(naphthalen-1-ylmethyl)-2H-pyrazolo[3,4-d]pyrimidine-4,6(5H,7H)-dione, D-GLUTAMIC ACID, GLUTAMATE RACEMASE | | Authors: | Basarab, G.S, Hill, P, Eyermann, C.J, Gowravaram, M, Kack, H, Osimoni, E. | | Deposit date: | 2012-07-10 | | Release date: | 2012-07-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Design of Inhibitors of Helicobacter Pylori Glutamate Racemase as Selective Antibacterial Agents: Incorporation of Imidazoles Onto a Core Pyrazolopyrimidinedione Scaffold to Improve Bioavailabilty.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
5PUF
 
 | | PanDDA analysis group deposition -- Crystal Structure of BRD1 after initial refinement with no ligand modelled (structure 193) | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 1, SODIUM ION | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-07 | | Release date: | 2017-03-29 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
4AW6
 
 | | Crystal structure of the human nuclear membrane zinc metalloprotease ZMPSTE24 (FACE1) | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CAAX PRENYL PROTEASE 1 HOMOLOG, ZINC ION | | Authors: | Pike, A.C.W, Dong, Y.Y, Quigley, A, Dong, L, Cooper, C.D.O, Chaikuad, A, Goubin, S, Shrestha, L, Li, Q, Mukhopadhyay, S, Yang, J, Xia, X, Shintre, C.A, Barr, A.J, Berridge, G, Chalk, R, Bray, J.E, von Delft, F, Bullock, A, Bountra, C, Arrowsmith, C.H, Edwards, A, Burgess-Brown, N, Carpenter, E.P. | | Deposit date: | 2012-05-31 | | Release date: | 2012-07-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | The Structural Basis of Zmpste24-Dependent Laminopathies.
Science, 339, 2013
|
|
5PUV
 
 | | PanDDA analysis group deposition -- Crystal Structure of BRD1 after initial refinement with no ligand modelled (structure 209) | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 1, SODIUM ION | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-07 | | Release date: | 2017-03-29 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
5PVA
 
 | | PanDDA analysis group deposition -- Crystal Structure of BRD1 after initial refinement with no ligand modelled (structure 224) | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 1, SODIUM ION | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-07 | | Release date: | 2017-03-29 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
5PVM
 
 | | PanDDA analysis group deposition -- Crystal Structure of BRD1 after initial refinement with no ligand modelled (structure 237) | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 1, SODIUM ION | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-07 | | Release date: | 2017-03-29 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
4N4E
 
 | | Thermolysin in complex with UBTLN58 | | Descriptor: | CALCIUM ION, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Krimmer, S.G, Heine, A, Klebe, G. | | Deposit date: | 2013-10-08 | | Release date: | 2014-04-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Methyl, Ethyl, Propyl, Butyl: Futile But Not for Water, as the Correlation of Structure and Thermodynamic Signature Shows in a Congeneric Series of Thermolysin Inhibitors.
Chemmedchem, 4, 2014
|
|
3MZ2
 
 | |
