7GND
 
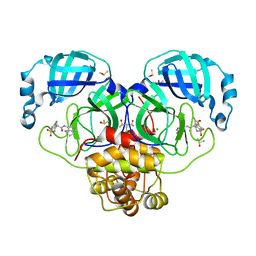 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with MIK-UNK-78dbf1b8-1 (Mpro-P2601) | | Descriptor: | (4S)-6-chloro-1,1-dioxo-N-(5,6,7,8-tetrahydroisoquinolin-4-yl)-1,2,3,4-tetrahydro-1lambda~6~,2-benzothiazine-4-carboxamide, 3C-like proteinase, CHLORIDE ION, ... | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.655 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
1WS1
 
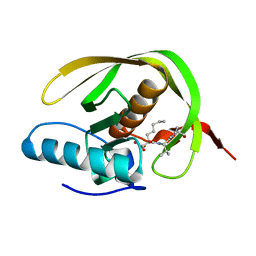 | |
3HWU
 
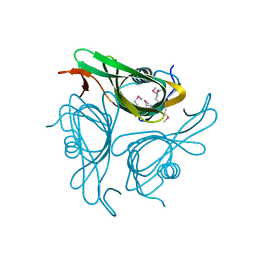 | |
4JQ8
 
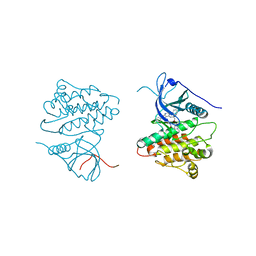 | | Crystal structure of EGFR kinase domain in complex with compound 4b | | Descriptor: | Epidermal growth factor receptor, N-[3-(4-{[(1S)-2-hydroxy-1-phenylethyl]amino}-6-phenylfuro[2,3-d]pyrimidin-5-yl)phenyl]-N~3~,N~3~-dimethyl-beta-alaninamide | | Authors: | Peng, Y.H, Wu, J.S. | | Deposit date: | 2013-03-20 | | Release date: | 2013-06-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | Protein Kinase Inhibitor Design by Targeting the Asp-Phe-Gly (DFG) Motif: The Role of the DFG Motif in the Design of Epidermal Growth Factor Receptor Inhibitors
J.Med.Chem., 56, 2013
|
|
4JAO
 
 | |
7GM0
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with MAT-POS-bfd29aac-1 (Mpro-P2070) | | Descriptor: | (4S)-6-chloro-2-[(1-cyanocyclopropyl)methanesulfonyl]-N-(1-methyl-1H-pyrazolo[4,3-c]pyridin-7-yl)-1,2,3,4-tetrahydroisoquinoline-4-carboxamide, 3C-like proteinase, CHLORIDE ION, ... | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
4JR3
 
 | | Crystal structure of EGFR kinase domain in complex with compound 3g | | Descriptor: | Epidermal growth factor receptor, N-[3-(4-{[(1S)-2-hydroxy-1-phenylethyl]amino}-6-phenylfuro[2,3-d]pyrimidin-5-yl)phenyl]acetamide | | Authors: | Peng, Y.H, Wu, J.S. | | Deposit date: | 2013-03-21 | | Release date: | 2013-06-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Protein Kinase Inhibitor Design by Targeting the Asp-Phe-Gly (DFG) Motif: The Role of the DFG Motif in the Design of Epidermal Growth Factor Receptor Inhibitors
J.Med.Chem., 56, 2013
|
|
3RNU
 
 | | Structural Basis of Cytosolic DNA Sensing by Innate Immune Receptors | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*GP*CP*CP*AP*TP*CP*AP*AP*AP*GP*AP*GP*AP*GP*AP*G)-3'), DNA (5'-D(*TP*CP*TP*CP*TP*CP*TP*TP*TP*GP*AP*TP*GP*GP*CP*C)-3'), ... | | Authors: | Jin, T.C, Xiao, T. | | Deposit date: | 2011-04-22 | | Release date: | 2012-04-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.502 Å) | | Cite: | Structures of the HIN Domain:DNA Complexes Reveal Ligand Binding and Activation Mechanisms of the AIM2 Inflammasome and IFI16 Receptor.
Immunity, 36, 2012
|
|
3MO5
 
 | | Human G9a-like (GLP, also known as EHMT1) in complex with inhibitor E72 | | Descriptor: | 7-[(5-aminopentyl)oxy]-N~4~-[1-(5-aminopentyl)piperidin-4-yl]-N~2~-[3-(dimethylamino)propyl]-6-methoxyquinazoline-2,4-diamine, Histone-lysine N-methyltransferase, H3 lysine-9 specific 5, ... | | Authors: | Chang, Y, Horton, J.R, Cheng, X. | | Deposit date: | 2010-04-22 | | Release date: | 2010-06-30 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Adding a lysine mimic in the design of potent inhibitors of histone lysine methyltransferases.
J.Mol.Biol., 400, 2010
|
|
2XZG
 
 | | Clathrin Terminal Domain Complexed with Pitstop 1 | | Descriptor: | 2-(4-AMINOBENZYL)-1,3-DIOXO-2,3-DIHYDRO-1H-BENZO[DE]ISOQUINOLINE-5-SULFONATE, ACETATE ION, CLATHRIN HEAVY CHAIN 1, ... | | Authors: | Bulut, H, Von Kleist, L, Saenger, W, Haucke, V. | | Deposit date: | 2010-11-25 | | Release date: | 2011-08-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Role of the Clathrin Terminal Domain in Regulating Coated Pit Dynamics Revealed by Small Molecule Inhibition.
Cell(Cambridge,Mass.), 146, 2011
|
|
3MG0
 
 | | Structure of yeast 20S proteasome with bortezomib | | Descriptor: | N-[(1R)-1-(DIHYDROXYBORYL)-3-METHYLBUTYL]-N-(PYRAZIN-2-YLCARBONYL)-L-PHENYLALANINAMIDE, Proteasome component C1, Proteasome component C11, ... | | Authors: | Sintchak, M.D. | | Deposit date: | 2010-04-05 | | Release date: | 2011-05-18 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Characterization of a new series of non-covalent proteasome inhibitors with exquisite potency and selectivity for the 20S beta5-subunit.
Biochem.J., 430, 2010
|
|
1UCA
 
 | | Crystal structure of the Ribonuclease MC1 from bitter gourd seeds complexed with 2'-UMP | | Descriptor: | PHOSPHORIC ACID MONO-[2-(2,4-DIOXO-3,4-DIHYDRO-2H-PYRIMIDIN-1-YL)-4-HYDROXY-5-HYDROXYMETHYL-TETRAHYDRO-FURAN-3-YL] ESTER, Ribonuclease MC | | Authors: | Suzuki, A, Yao, M, Tanaka, I, Numata, T, Kikukawa, S, Yamasaki, N, Kimura, M. | | Deposit date: | 2003-04-10 | | Release date: | 2003-04-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Crystal structures of the ribonuclease MC1 from bitter gourd seeds, complexed with 2'-UMP or 3'-UMP, reveal structural basis for uridine specificity
Biochem.Biophys.Res.Commun., 275, 2000
|
|
3BGM
 
 | | Crystal Structure of PKD2 Phosphopeptide Bound to Human Class I MHC HLA-A2 | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, HLA class I histocompatibility antigen, ... | | Authors: | Mohammed, F, Cobbold, M, Zarling, A.L, Salim, M, Barrett-Wilt, G.A, Shabanowitz, J, Hunt, D.F, Engelhard, V.H, Willcox, B.E. | | Deposit date: | 2007-11-27 | | Release date: | 2008-10-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Phosphorylation-dependent interaction between antigenic peptides and MHC class I: a molecular basis for the presentation of transformed self
Nat.Immunol., 9, 2008
|
|
3BHH
 
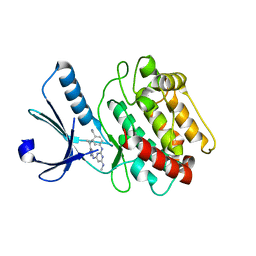 | | Crystal structure of human calcium/calmodulin-dependent protein kinase IIB isoform 1 (CAMK2B) | | Descriptor: | Calcium/calmodulin-dependent protein kinase type II beta chain, [4-({4-[(5-cyclopropyl-1H-pyrazol-3-yl)amino]-6-(methylamino)pyrimidin-2-yl}amino)phenyl]acetonitrile | | Authors: | Filippakopoulos, P, Rellos, P, Niesen, F, Burgess, N, Bullock, A, Berridge, G, Pike, A.C.W, Ugochukwu, E, Pilka, E.S, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-11-28 | | Release date: | 2007-12-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of Human Calcium/Calmodulin-Dependent Protein Kinase IIB Isoform 1 (CAMK2B).
To be Published
|
|
4OYM
 
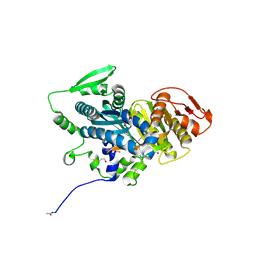 | | Human solAC Complexed with (4-Amino-furazan-3-yl)-(3-methoxy-phenyl)-methanone | | Descriptor: | (4-azanyl-1,2,5-oxadiazol-3-yl)-(3-methoxyphenyl)methanone, Adenylate cyclase type 10, GLYCEROL | | Authors: | Vinkovic, M. | | Deposit date: | 2014-02-12 | | Release date: | 2014-04-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of human soluble adenylate cyclase reveals a distinct, highly flexible allosteric bicarbonate binding pocket.
Chemmedchem, 9, 2014
|
|
7FKQ
 
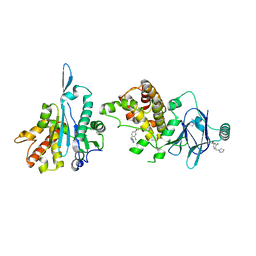 | | PanDDA analysis group deposition -- Aar2/RNaseH in complex with fragment P04E09 from the F2X-Universal Library | | Descriptor: | 1-[(2-chloro-4-methoxyphenyl)methyl]-1H-1,2,4-triazole, A1 cistron-splicing factor AAR2, Pre-mRNA-splicing factor 8 | | Authors: | Barthel, T, Wollenhaupt, J, Lima, G.M.A, Wahl, M.C, Weiss, M.S. | | Deposit date: | 2022-08-26 | | Release date: | 2022-11-02 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Large-Scale Crystallographic Fragment Screening Expedites Compound Optimization and Identifies Putative Protein-Protein Interaction Sites.
J.Med.Chem., 65, 2022
|
|
3HOI
 
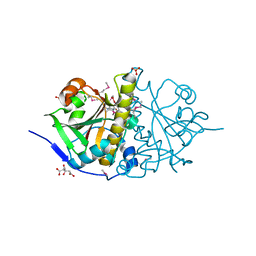 | |
5PAO
 
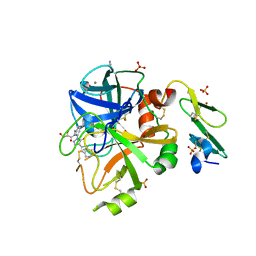 | | Crystal Structure of Factor VIIa in complex with (2S)-2,3-dihydroxy-N-[[3-[5-hydroxy-4-(1H-pyrrolo[3,2-c]pyridin-2-yl)pyrazol-1-yl]phenyl]methyl]propanamide;hydrobromide | | Descriptor: | (2S)-2,3-dihydroxy-N-[[3-[5-hydroxy-4-(1H-pyrrolo[3,2-c]pyridin-2-yl)pyrazol-1-yl]phenyl]methyl]propanamide, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Stihle, M, Mayweg, A, Roever, S, Rudolph, M.G. | | Deposit date: | 2016-11-10 | | Release date: | 2017-06-21 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal Structure of a Factor VIIa complex
To be published
|
|
3APP
 
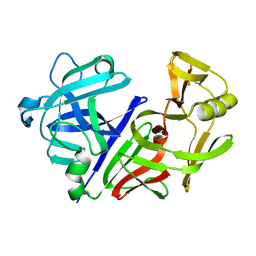 | |
3ZPI
 
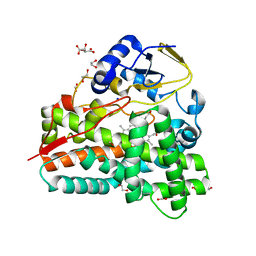 | | PikC D50N mutant in P21 space group | | Descriptor: | 1,2-ETHANEDIOL, CYTOCHROME P450 HYDROXYLASE PIKC, FORMIC ACID, ... | | Authors: | Podust, L.M. | | Deposit date: | 2013-02-27 | | Release date: | 2014-03-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Recognition of Synthetic Substrates by P450 Pikc
To be Published
|
|
5TBC
 
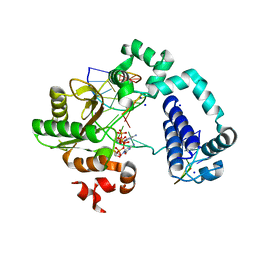 | |
4JWT
 
 | |
3MMX
 
 | | Bacillus anthracis NadD (baNadD) in complex with compound 1_02_3 | | Descriptor: | CITRIC ACID, DIMETHYL SULFOXIDE, POTASSIUM ION, ... | | Authors: | Huang, N, Zhang, H. | | Deposit date: | 2010-04-20 | | Release date: | 2010-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.552 Å) | | Cite: | Complexes of bacterial nicotinate mononucleotide adenylyltransferase with inhibitors: implication for structure-based drug design and improvement.
J.Med.Chem., 53, 2010
|
|
3QM5
 
 | | Blackfin tuna oxy-myoglobin, atomic resolution | | Descriptor: | 1,2-ETHANEDIOL, Myoglobin, OXYGEN MOLECULE, ... | | Authors: | de Serrano, V.S, Rodriguez, M.M, Schreiter, E.R. | | Deposit date: | 2011-02-03 | | Release date: | 2012-02-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (0.91 Å) | | Cite: | High Resolution Structures of Blackfin Tuna Myoglobin
To be Published
|
|
3BRJ
 
 | | Crystal structure of mannitol operon repressor (MtlR) from Vibrio parahaemolyticus RIMD 2210633 | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Mannitol operon repressor | | Authors: | Tan, K, Zhou, M, Moy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-12-21 | | Release date: | 2008-01-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | The mannitol operon repressor MtlR belongs to a new class of transcription regulators in bacteria.
J.Biol.Chem., 284, 2009
|
|
