1EVN
 
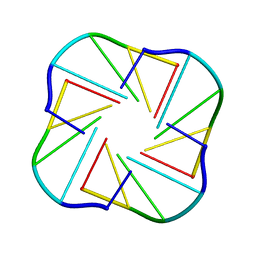 | | NMR OBSERVATION OF A-TETRAD | | Descriptor: | DNA (5'-D(*AP*GP*GP*GP*T)-3') | | Authors: | Patel, P.K, Koti, A.S.R, Hosur, R.V. | | Deposit date: | 2000-04-20 | | Release date: | 2000-05-22 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR studies on truncated sequences of human telomeric DNA: observation of a novel A-tetrad.
Nucleic Acids Res., 27, 1999
|
|
1DV0
 
 | |
3AL2
 
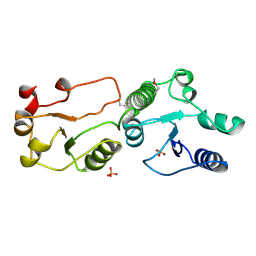 | | Crystal Structure of TopBP1 BRCT7/8 | | Descriptor: | DNA topoisomerase 2-binding protein 1, SULFATE ION | | Authors: | Leung, C.C, Glover, J.N. | | Deposit date: | 2010-07-22 | | Release date: | 2010-12-01 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular basis of BACH1/FANCJ recognition by TopBP1 in DNA replication checkpoint control
J.Biol.Chem., 286, 2011
|
|
3AL3
 
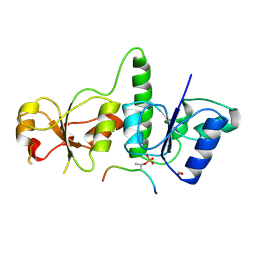 | | Crystal Structure of TopBP1 BRCT7/8-BACH1 peptide complex | | Descriptor: | DNA topoisomerase 2-binding protein 1, FORMIC ACID, Peptide of Fanconi anemia group J protein | | Authors: | Leung, C.C, Glover, J.N. | | Deposit date: | 2010-07-22 | | Release date: | 2010-12-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Molecular basis of BACH1/FANCJ recognition by TopBP1 in DNA replication checkpoint control
J.Biol.Chem., 286, 2011
|
|
3CWV
 
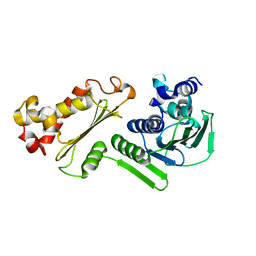 | | Crystal structure of B-subunit of the DNA gyrase from Myxococcus xanthus | | Descriptor: | DNA gyrase, B subunit, truncated | | Authors: | Ramagopal, U.A, Toro, R, Meyer, A.J, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-04-22 | | Release date: | 2008-05-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of B-subunit of the DNA gyrase from Myxococcus xanthus.
To be published
|
|
5WTU
 
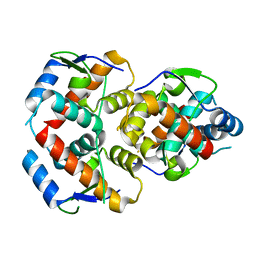 | |
1EVM
 
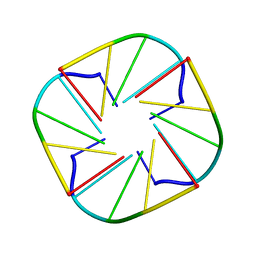 | | NMR OBSERVATION OF A-TETRAD | | Descriptor: | DNA (5'-D(*AP*GP*GP*GP*T)-3') | | Authors: | Patel, P.K, Koti, A.S.R, Hosur, R.V. | | Deposit date: | 2000-04-20 | | Release date: | 2000-05-22 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR studies on truncated sequences of human telomeric DNA: observation of a novel A-tetrad.
Nucleic Acids Res., 27, 1999
|
|
2E2W
 
 | |
2M6V
 
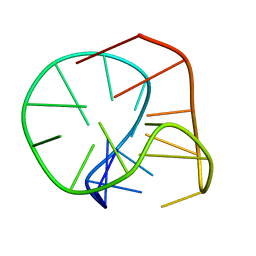 | |
2KTF
 
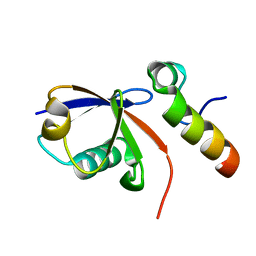 | |
1EHV
 
 | |
2L0G
 
 | |
2L0F
 
 | |
3QKU
 
 | | Mre11 Rad50 binding domain in complex with Rad50 and AMP-PNP | | Descriptor: | DNA double-strand break repair protein mre11, DNA double-strand break repair rad50 ATPase, MAGNESIUM ION, ... | | Authors: | Williams, G.J, Williams, R.S, Arvai, A, Moncalian, G, Tainer, J.A. | | Deposit date: | 2011-02-01 | | Release date: | 2011-03-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | ABC ATPase signature helices in Rad50 link nucleotide state to Mre11 interface for DNA repair.
Nat.Struct.Mol.Biol., 18, 2011
|
|
3QKR
 
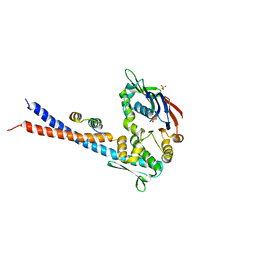 | | Mre11 Rad50 binding domain bound to Rad50 | | Descriptor: | DNA double-strand break repair protein mre11, DNA double-strand break repair rad50 ATPase, PHOSPHATE ION | | Authors: | Williams, G.J, Williams, R.S, Arvai, A, Moncalian, G, Tainer, J.A. | | Deposit date: | 2011-02-01 | | Release date: | 2011-03-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | ABC ATPase signature helices in Rad50 link nucleotide state to Mre11 interface for DNA repair.
Nat.Struct.Mol.Biol., 18, 2011
|
|
386D
 
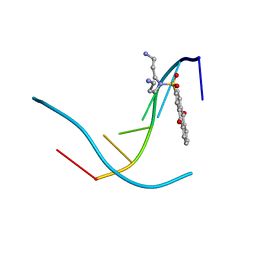 | | THREE-DIMENSIONAL STRUCTURE AND REACTIVITY OF A PHOTOCHEMICAL CLEAVAGE AGENT BOUND TO DNA | | Descriptor: | DNA (5'-D(*CP*GP*TP*AP*CP*G)-3'), N,N-BIS(3-AMINOPROPYL)-2-ANTHRAQUINONESULFONAMIDE | | Authors: | Gasper, S.M, Armitage, B, Shui, X, Hu, G.G, Yu, C, Schuster, G, Williams, L.D. | | Deposit date: | 1998-03-11 | | Release date: | 1998-03-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Three-Dimensional Structure and Reactivity of a Photochemical Cleavage Agent Bound to DNA
J.Am.Chem.Soc., 120, 1998
|
|
3QKS
 
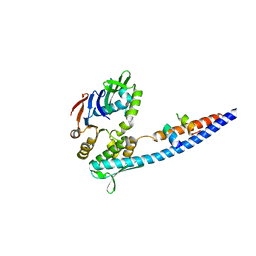 | | Mre11 Rad50 binding domain bound to Rad50 | | Descriptor: | DNA double-strand break repair protein mre11, DNA double-strand break repair rad50 ATPase | | Authors: | Williams, G.J, Williams, R.S, Arvai, A, Moncalian, G, Tainer, J.A. | | Deposit date: | 2011-02-01 | | Release date: | 2011-03-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | ABC ATPase signature helices in Rad50 link nucleotide state to Mre11 interface for DNA repair.
Nat.Struct.Mol.Biol., 18, 2011
|
|
240D
 
 | |
2QS6
 
 | | Structure of a Hoogsteen antiparallel duplex with extra-helical thymines | | Descriptor: | DNA (5'-D(*DAP*DTP*DAP*DTP*DAP*DTP*DCP*DT)-3') | | Authors: | Pous, J, Urpi, L, Subirana, J.A, Gouyette, C, Navaza, J, Campos, J.L. | | Deposit date: | 2007-07-30 | | Release date: | 2008-03-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.08 Å) | | Cite: | Stabilization by extra-helical thymines of a DNA duplex with Hoogsteen base pairs.
J.Am.Chem.Soc., 130, 2008
|
|
6FRG
 
 | | Crystal structure of G-1F mutant of Ssp DnaB Mini-Intein variant M86 | | Descriptor: | DI(HYDROXYETHYL)ETHER, PENTAETHYLENE GLYCOL, Replicative DNA helicase, ... | | Authors: | Popp, M.A, Blankenfeldt, W, Friedel, K, Mootz, H.D. | | Deposit date: | 2018-02-15 | | Release date: | 2019-01-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.535 Å) | | Cite: | A functional interplay between intein and extein sequences in protein splicing compensates for the essential block B histidine.
Chem Sci, 10, 2019
|
|
2FAQ
 
 | | Crystal Structure of Pseudomonas aeruginosa LigD polymerase domain with ATP and Manganese | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MANGANESE (II) ION, SULFATE ION, ... | | Authors: | Zhu, H, Nandakumar, J, Aniukwu, J, Wang, L.K, Glickman, M.S, Lima, C.D, Shuman, S. | | Deposit date: | 2005-12-07 | | Release date: | 2006-05-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Atomic structure and nonhomologous end-joining function of the polymerase component of bacterial DNA ligase D
Proc.Natl.Acad.Sci.USA, 103, 2006
|
|
2FHD
 
 | | Crystal structure of Crb2 tandem tudor domains | | Descriptor: | DNA repair protein rhp9/CRB2, PHOSPHATE ION | | Authors: | Lee, J, Botuyan, M.V, Thompson, J.R, Mer, G. | | Deposit date: | 2005-12-23 | | Release date: | 2007-01-02 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for the methylation state-specific recognition of histone H4-K20 by 53BP1 and Crb2 in DNA repair.
Cell(Cambridge,Mass.), 127, 2006
|
|
2FAO
 
 | | Crystal Structure of Pseudomonas aeruginosa LigD polymerase domain | | Descriptor: | SULFATE ION, probable ATP-dependent DNA ligase | | Authors: | Zhu, H, Nandakumar, J, Aniukwu, J, Wang, L.K, Glickman, M.S, Lima, C.D, Shuman, S. | | Deposit date: | 2005-12-07 | | Release date: | 2006-05-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Atomic structure and nonhomologous end-joining function of the polymerase component of bacterial DNA ligase D
Proc.Natl.Acad.Sci.USA, 103, 2006
|
|
2FAR
 
 | | Crystal Structure of Pseudomonas aeruginosa LigD polymerase domain with dATP and Manganese | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, MANGANESE (II) ION, SULFATE ION, ... | | Authors: | Zhu, H, Nandakumar, J, Aniukwu, J, Wang, L.K, Glickman, M.S, Lima, C.D, Shuman, S. | | Deposit date: | 2005-12-07 | | Release date: | 2006-05-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Atomic structure and nonhomologous end-joining function of the polymerase component of bacterial DNA ligase D
Proc.Natl.Acad.Sci.USA, 103, 2006
|
|
1KOY
 
 | | NMR structure of DFF-C domain | | Descriptor: | DNA fragmentation factor alpha subunit | | Authors: | Fukushima, K, Kikuchi, J, Koshiba, S, Kigawa, T, Kuroda, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2001-12-25 | | Release date: | 2002-09-04 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the DFF-C domain of DFF45/ICAD. A structural basis for the regulation of apoptotic DNA fragmentation.
J.Mol.Biol., 321, 2002
|
|
