1PJE
 
 | | Structure of the channel-forming trans-membrane domain of Virus protein "u"(Vpu) from HIV-1 | | Descriptor: | VPU protein | | Authors: | Park, S.H, Mrse, A.A, Nevzorov, A.A, Mesleh, M.F, Oblatt-Montal, M, Montal, M, Opella, S.J. | | Deposit date: | 2003-06-02 | | Release date: | 2003-10-14 | | Last modified: | 2024-05-22 | | Method: | SOLID-STATE NMR | | Cite: | Three-dimensional structure of the channel-forming trans-membrane domain of Virus protein "u" (Vpu) from HIV-1
J.Mol.Biol., 333, 2003
|
|
5W54
 
 | |
6IZG
 
 | |
6M6F
 
 | |
4CH1
 
 | |
4CIO
 
 | |
1Q2Z
 
 | | The 3D solution structure of the C-terminal region of Ku86 | | Descriptor: | ATP-dependent DNA helicase II, 80 kDa subunit | | Authors: | Harris, R, Esposito, D, Sankar, A, Maman, J.D, Hinks, J.A, Pearl, L.H, Driscoll, P.C. | | Deposit date: | 2003-07-28 | | Release date: | 2004-01-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The 3D Solution Structure of the C-terminal Region of Ku86 (Ku86CTR)
J.Mol.Biol., 335, 2004
|
|
6GQ9
 
 | |
1N8M
 
 | | Solution structure of Pi4, a four disulfide bridged scorpion toxin active on potassium channels | | Descriptor: | Potassium channel blocking toxin 4 | | Authors: | Guijarro, J.I, M'Barek, S, Olamendi-Portugal, T, Gomez-Lagunas, F, Garnier, D, Rochat, H, Possani, L.D, Sabatier, J.M, Delepierre, M. | | Deposit date: | 2002-11-21 | | Release date: | 2003-09-02 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Pi4, a short four-disulfide-bridged scorpion toxin specific of potassium channels.
Protein Sci., 12, 2003
|
|
8A6M
 
 | |
6MUN
 
 | | Structure of hRpn10 bound to UBQLN2 UBL | | Descriptor: | 26S proteasome non-ATPase regulatory subunit 4, Ubiquilin-2 | | Authors: | Chen, X, Walters, K.J. | | Deposit date: | 2018-10-23 | | Release date: | 2019-09-04 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of hRpn10 Bound to UBQLN2 UBL Illustrates Basis for Complementarity between Shuttle Factors and Substrates at the Proteasome.
J.Mol.Biol., 431, 2019
|
|
6MXQ
 
 | |
3ZPD
 
 | |
6NOA
 
 | |
5Y4B
 
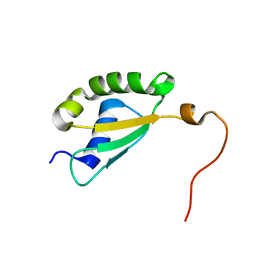 | | Solution structure of yeast Fra2 | | Descriptor: | BolA-like protein 2 | | Authors: | Tang, Y.J, Chi, C.B, Zhang, J.H, Dai, Y.N, Abdalla, M, Chen, Y.X, Zhou, C.Z. | | Deposit date: | 2017-08-03 | | Release date: | 2018-03-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural and Biochemical Insights into the Multiple Functions of Yeast Grx3.
J. Mol. Biol., 430, 2018
|
|
6H0Q
 
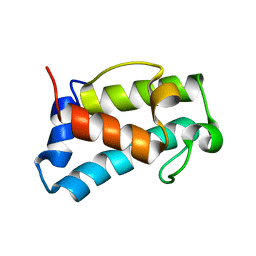 | | B1-type ACP domain from module 7 of MLSB | | Descriptor: | Type I modular polyketide synthase | | Authors: | Moretto, L, Heylen, R, Holroyd, N, Vance, S, Broadhurst, R.W. | | Deposit date: | 2018-07-10 | | Release date: | 2019-03-06 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Modular type I polyketide synthase acyl carrier protein domains share a common N-terminally extended fold.
Sci Rep, 9, 2019
|
|
6H0J
 
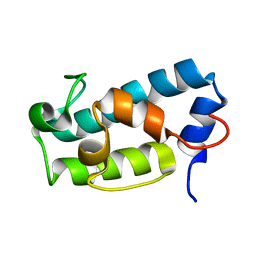 | | A1-type ACP domain from module 5 of MLSA1 | | Descriptor: | Type I modular polyketide synthase | | Authors: | Moretto, L, Heylen, R, Holroyd, N, Vance, S, Broadhurst, R.W. | | Deposit date: | 2018-07-09 | | Release date: | 2019-03-06 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Modular type I polyketide synthase acyl carrier protein domains share a common N-terminally extended fold.
Sci Rep, 9, 2019
|
|
6NW8
 
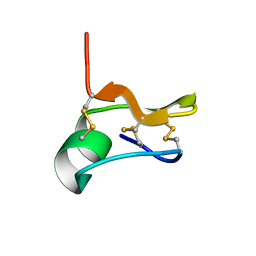 | | SOLUTION STRUCTURE OF CN29, A TOXIN FROM CENTRUROIDES NOXIUS SCORPION VENOM | | Descriptor: | Cn29 | | Authors: | Delepierre, M, Gurrola, G.B, Possani, L.D, Guijarro, J.I. | | Deposit date: | 2019-02-06 | | Release date: | 2019-07-03 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Cn29, a novel orphan peptide found in the venom of the scorpion Centruroides noxius: Structure and function.
Toxicon, 167, 2019
|
|
6NZN
 
 | | Dimer-of-dimer amyloid fibril structure of glucagon | | Descriptor: | Glucagon | | Authors: | Gelenter, M.D, Smith, K.J, Liao, S.Y, Mandala, V.S, Dregni, A.J, Lamm, M.S, Tian, Y, Wei, X, Pochan, D.J, Tucker, T.J, Su, Y, Hong, M. | | Deposit date: | 2019-02-14 | | Release date: | 2019-06-05 | | Last modified: | 2024-05-15 | | Method: | SOLID-STATE NMR | | Cite: | The peptide hormone glucagon forms amyloid fibrils with two coexisting beta-strand conformations.
Nat.Struct.Mol.Biol., 26, 2019
|
|
5X6T
 
 | |
5VWL
 
 | |
6P6B
 
 | |
6P6C
 
 | |
5YJ5
 
 | |
6PQG
 
 | |
