7I4M
 
 | | Group deposition of Coxsackievirus A16 (G-10) 2A protease in complex with inhibitors from the ASAP AViDD centre -- Crystal structure of Coxsackievirus A16 (G-10) 2A protease in complex with ASAP-0036643-001 (A71EV2A-x4715) | | Descriptor: | 1-(3-hydroxyazetidin-1-yl)-2-[2-(methylsulfanyl)phenyl]ethan-1-one, DIMETHYL SULFOXIDE, Protease 2A, ... | | Authors: | Lithgo, R.M, Fairhead, M, Koekemoer, L, Balcomb, B.H, Capkin, E, Chandran, A.V, Golding, M, Godoy, A.S, Aschenbrenner, J.C, Marples, P.G, Ni, X, Thompson, W, Tomlinson, C.W.E, Wild, C, Winokan, M, Xavier, M.-A.E, Kenton, N, Tucker, J, DiPoto, M, Lee, A, Fearon, D, von Delft, F. | | Deposit date: | 2025-03-12 | | Release date: | 2025-04-02 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | Group deposition of Coxsackievirus A16 (G-10) 2A protease in complex with inhibitors from the ASAP AViDD centre
To Be Published
|
|
4R0Q
 
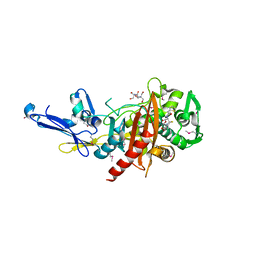 | | Structure of a putative peptidoglycan glycosyltransferase from Atopobium parvulum in complex with cephalothin | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CEPHALOTHIN GROUP, Peptidoglycan glycosyltransferase, ... | | Authors: | Filippova, E.V, Minasov, G, Kiryukhina, O, Clancy, S, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-08-01 | | Release date: | 2014-08-27 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of a putative peptidoglycan glycosyltransferase from Atopobium parvulum in complex with cephalothin
To be Published
|
|
7I5A
 
 | | Group deposition of Coxsackievirus A16 (G-10) 2A protease in complex with inhibitors from the ASAP AViDD centre -- Crystal structure of Coxsackievirus A16 (G-10) 2A protease in complex with ASAP-0036771-001 (A71EV2A-x4809) | | Descriptor: | 1-[(3R)-3-(2-hydroxyethyl)piperidin-1-yl]-2-[2-(methylsulfanyl)phenyl]ethan-1-one, Protease 2A, ZINC ION | | Authors: | Lithgo, R.M, Fairhead, M, Koekemoer, L, Balcomb, B.H, Capkin, E, Chandran, A.V, Golding, M, Godoy, A.S, Aschenbrenner, J.C, Marples, P.G, Ni, X, Thompson, W, Tomlinson, C.W.E, Wild, C, Winokan, M, Xavier, M.-A.E, Kenton, N, Tucker, J, DiPoto, M, Lee, A, Fearon, D, von Delft, F. | | Deposit date: | 2025-03-12 | | Release date: | 2025-04-02 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.449 Å) | | Cite: | Group deposition of Coxsackievirus A16 (G-10) 2A protease in complex with inhibitors from the ASAP AViDD centre
To Be Published
|
|
4NEW
 
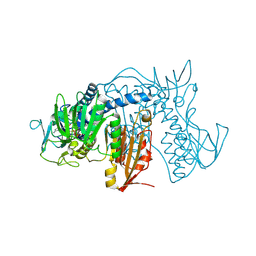 | | Crystal structure of Trypanothione Reductase from Trypanosoma cruzi in complex with inhibitor EP127 (5-{5-[1-(PYRROLIDIN-1-YL)CYCLOHEXYL]-1,3-THIAZOL-2-YL}-1H-INDOLE) | | Descriptor: | 5-{5-[1-(pyrrolidin-1-yl)cyclohexyl]-1,3-thiazol-2-yl}-1H-indole, FLAVIN-ADENINE DINUCLEOTIDE, Trypanothione reductase, ... | | Authors: | Persch, E, Bryson, S, Pai, E.F, Krauth-Siegel, R.L, Diederich, F. | | Deposit date: | 2013-10-30 | | Release date: | 2014-05-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Binding to large enzyme pockets: small-molecule inhibitors of trypanothione reductase.
Chemmedchem, 9, 2014
|
|
4JEX
 
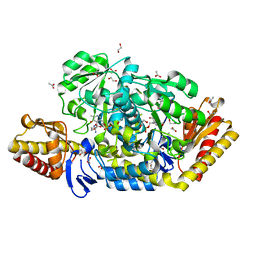 | | Y21K mutant of N-acetylornithine aminotransferase complexed with L-canaline | | Descriptor: | (2S)-2-azanyl-4-[(E)-[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]methylideneamino]oxy-butanoic acid, 1,2-ETHANEDIOL, ACETATE ION, ... | | Authors: | Bisht, S, Bharath, S.R, Murthy, M.R.N. | | Deposit date: | 2013-02-27 | | Release date: | 2014-03-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Conformational transitions, ligand specificity and catalysis in N-acetylornithine aminotransferase: Implications on drug designing and rational enzyme engineering in omega aminotransferases
To be Published
|
|
7CZM
 
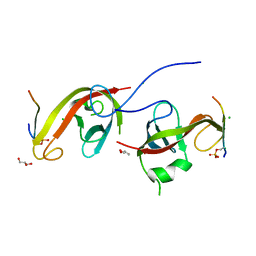 | | Crystal structure of FIP200 Claw/p-OPtineurin LIR complex | | Descriptor: | CHLORIDE ION, GLYCEROL, Optineurin LIR, ... | | Authors: | Zhou, Z.X, Pan, L.F. | | Deposit date: | 2020-09-09 | | Release date: | 2021-03-31 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Phosphorylation regulates the binding of autophagy receptors to FIP200 Claw domain for selective autophagy initiation.
Nat Commun, 12, 2021
|
|
5S67
 
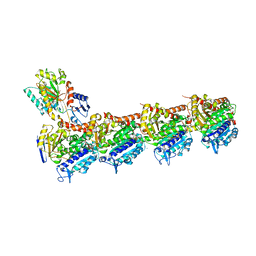 | | Tubulin-Z1896597864-complex | | Descriptor: | 1-(6-methoxypyridin-2-yl)-N-methylmethanamine, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Muehlethaler, T, Gioia, D, Prota, A.E, Sharpe, M.E, Cavalli, A, Steinmetz, M.O. | | Deposit date: | 2020-11-08 | | Release date: | 2021-06-30 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Comprehensive Analysis of Binding Sites in Tubulin.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
4BCF
 
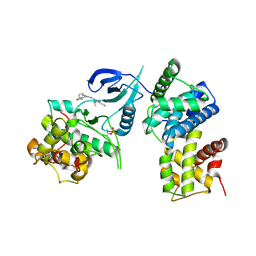 | | Structure of CDK9 in complex with cyclin T and a 2-amino-4-heteroaryl- pyrimidine inhibitor | | Descriptor: | 2-[[3-(4-ethanoyl-1,4-diazepan-1-yl)phenyl]amino]-4-[4-methyl-2-(methylamino)-1,3-thiazol-5-yl]pyrimidine-5-carbonitrile, CYCLIN-DEPENDENT KINASE 9, CYCLIN-T1 | | Authors: | Hole, A.J, Baumli, S, Wang, S, Endicott, J.A, Noble, M.E.M. | | Deposit date: | 2012-10-02 | | Release date: | 2013-01-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.011 Å) | | Cite: | Comparative Structural and Functional Studies of 4-(Thiazol- 5-Yl)-2-(Phenylamino)Pyrimidine-5-Carbonitrile Cdk9 Inhibitors Suggest the Basis for Isotype Selectivity.
J.Med.Chem., 56, 2013
|
|
4JEZ
 
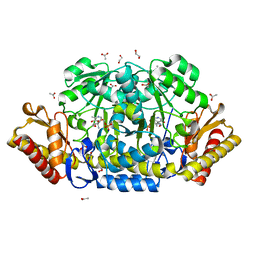 | | N79R mutant of N-acetylornithine aminotransferase complexed with L-canaline | | Descriptor: | (2S)-2-azanyl-4-[(E)-[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]methylideneamino]oxy-butanoic acid, 1,2-ETHANEDIOL, ACETATE ION, ... | | Authors: | Bisht, S, Bharath, S.R, Murthy, M.R.N. | | Deposit date: | 2013-02-27 | | Release date: | 2014-03-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Conformational transitions, ligand specificity and catalysis in N-acetylornithine aminotransferase: Implications on drug designing and rational enzyme engineering in omega aminotransferases
To be Published
|
|
5QIS
 
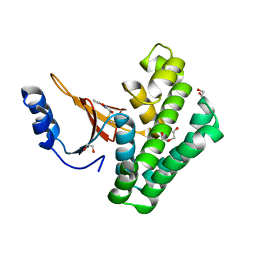 | | Covalent fragment group deposition -- Crystal Structure of OUTB2 in complex with PCM-0102500 | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, N-(5-methyl-1,2-oxazol-3-yl)acetamide, ... | | Authors: | Sethi, R, Douangamath, A, Resnick, E, Bradley, A.R, Collins, P, Brandao-Neto, J, Talon, R, Krojer, T, Bountra, C, Arrowsmith, C.H, Edwards, A, London, N, von Delft, F. | | Deposit date: | 2018-08-10 | | Release date: | 2019-12-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Covalent fragment group deposition
To Be Published
|
|
8DL2
 
 | | BoGH13ASus from Bacteroides ovatus bound to acarbose | | Descriptor: | 1,2-ETHANEDIOL, 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, ... | | Authors: | Brown, H.A, DeVeaux, A.L, Koropatkin, N.M. | | Deposit date: | 2022-07-06 | | Release date: | 2023-05-24 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | BoGH13A Sus from Bacteroides ovatus represents a novel alpha-amylase used for Bacteroides starch breakdown in the human gut.
Cell.Mol.Life Sci., 80, 2023
|
|
4B7T
 
 | | Glycogen Synthase Kinase 3 Beta complexed with Axin Peptide and Leucettine L4 | | Descriptor: | (5Z)-5-(1,3-benzodioxol-5-ylmethylidene)-3-methyl-2-(propan-2-ylamino)imidazol-4-one, AXIN-1, GLYCOGEN SYNTHASE KINASE-3 BETA | | Authors: | Oberholzer, A.E, Pearl, L.H. | | Deposit date: | 2012-08-22 | | Release date: | 2013-01-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.772 Å) | | Cite: | Selectivity, Cocrystal Structures, and Neuroprotective Properties of Leucettines, a Family of Protein Kinase Inhibitors Derived from the Marine Sponge Alkaloid Leucettamine B.
J.Med.Chem., 55, 2012
|
|
3QXS
 
 | | Crystal structure of dethiobiotin synthetase (BioD) from Helicobacter pylori complexed with ANP | | Descriptor: | 1,2-ETHANEDIOL, Dethiobiotin synthetase, MAGNESIUM ION, ... | | Authors: | Klimecka, M.M, Porebski, P.J, Chruszcz, M, Jablonska, K, Murzyn, K, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-03-02 | | Release date: | 2011-03-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural characterization of Helicobacter pylori dethiobiotin synthetase reveals differences between family members.
Febs J., 279, 2012
|
|
3IW8
 
 | | Structure of Inactive Human p38 MAP Kinase in Complex with a Thiazole-Urea | | Descriptor: | 1-{4-[(1S)-1-amino-2-(benzyloxy)ethyl]-1,3-thiazol-2-yl}-3-(3-chloro-4-fluorophenyl)urea, Mitogen-activated protein kinase 14, octyl beta-D-glucopyranoside | | Authors: | Gruetter, C, Simard, J.R, Rauh, D. | | Deposit date: | 2009-09-02 | | Release date: | 2009-11-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | High-Throughput Screening To Identify Inhibitors Which Stabilize Inactive Kinase Conformations in p38alpha
J.Am.Chem.Soc., 131, 2009
|
|
7DFL
 
 | | Cryo-EM structure of histamine H1 receptor Gq complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(q) subunit alpha, ... | | Authors: | He, Y, Xia, R, Wang, N, Xu, Z. | | Deposit date: | 2020-11-09 | | Release date: | 2021-03-31 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structure of the human histamine H 1 receptor/G q complex.
Nat Commun, 12, 2021
|
|
1UOT
 
 | | HUMAN CD55 DOMAINS 3 & 4 | | Descriptor: | COMPLEMENT DECAY-ACCELERATING FACTOR | | Authors: | Williams, P, Chaudhry, Y, Goodfellow, I.G, Billington, J, Spiller, B, Evans, D.J, Lea, S.M. | | Deposit date: | 2003-09-23 | | Release date: | 2003-09-25 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Mapping Cd55 Function. The Structure of Two Pathogen-Binding Domains at 1.7 A
J.Biol.Chem., 278, 2003
|
|
2Y6O
 
 | |
5S63
 
 | | Tubulin-Z2241115980-complex | | Descriptor: | 1-[(furan-2-yl)methyl]-4-(methylsulfonyl)piperazine, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Muehlethaler, T, Gioia, D, Prota, A.E, Sharpe, M.E, Cavalli, A, Steinmetz, M.O. | | Deposit date: | 2020-11-08 | | Release date: | 2021-06-30 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Comprehensive Analysis of Binding Sites in Tubulin.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
4B1G
 
 | | Structure of unliganded human PARG catalytic domain | | Descriptor: | (2S,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, POLY(ADP-RIBOSE) GLYCOHYDROLASE, SULFATE ION | | Authors: | Brassington, C, Ellston, J, Hassall, G, Holdgate, G, McAlister, M, Overman, R, Smith, G, Tucker, J.A, Watson, M. | | Deposit date: | 2012-07-10 | | Release date: | 2012-12-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structures of the Human Poly (Adp-Ribose) Glycohydrolase Catalytic Domain Confirm Catalytic Mechanism and Explain Inhibition by Adp-Hpd Derivatives.
Plos One, 7, 2012
|
|
2Y8E
 
 | |
4BA1
 
 | |
7I4Y
 
 | | Group deposition of Coxsackievirus A16 (G-10) 2A protease in complex with inhibitors from the ASAP AViDD centre -- Crystal structure of Coxsackievirus A16 (G-10) 2A protease in complex with ASAP-0036788-001 (A71EV2A-x4752) | | Descriptor: | 1-(4-hydroxy-7,8-dihydro-1,6-naphthyridin-6(5H)-yl)-2-[2-(methylsulfanyl)phenyl]ethan-1-one, DIMETHYL SULFOXIDE, Protease 2A, ... | | Authors: | Lithgo, R.M, Fairhead, M, Koekemoer, L, Balcomb, B.H, Capkin, E, Chandran, A.V, Golding, M, Godoy, A.S, Aschenbrenner, J.C, Marples, P.G, Ni, X, Thompson, W, Tomlinson, C.W.E, Wild, C, Winokan, M, Xavier, M.-A.E, Kenton, N, Tucker, J, DiPoto, M, Lee, A, Fearon, D, von Delft, F. | | Deposit date: | 2025-03-12 | | Release date: | 2025-04-02 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.631 Å) | | Cite: | Group deposition of Coxsackievirus A16 (G-10) 2A protease in complex with inhibitors from the ASAP AViDD centre
To Be Published
|
|
1UV5
 
 | | GLYCOGEN SYNTHASE KINASE 3 BETA COMPLEXED WITH 6-BROMOINDIRUBIN-3'-OXIME | | Descriptor: | 6-BROMOINDIRUBIN-3'-OXIME, CHLORIDE ION, COBALT (II) ION, ... | | Authors: | Dajani, R, Pearl, L.H, Roe, S.M. | | Deposit date: | 2004-01-14 | | Release date: | 2004-01-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Gsk-3-Selective Inhibitors Derived from Tyrian Purple Indurubins
Chem.Biol., 10, 2003
|
|
3LC5
 
 | | Selective Benzothiophine Inhibitors of Factor IXa | | Descriptor: | 1-{4-[(R)-phenyl(3-phenyl-1,2,4-oxadiazol-5-yl)methoxy]-1-benzothiophen-2-yl}methanediamine, CALCIUM ION, Coagulation factor IX | | Authors: | Wang, S, Beck, R. | | Deposit date: | 2010-01-09 | | Release date: | 2010-02-23 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Structure Based Drug Design: Development of Potent and Selective Factor IXa (FIXa) Inhibitors.
J.Med.Chem., 53, 2010
|
|
2OC2
 
 | | Structure of testis ACE with RXPA380 | | Descriptor: | Angiotensin-converting enzyme, somatic isoform, CHLORIDE ION, ... | | Authors: | Corradi, H.R, Anthony, C.S, Schwager, S.L, Redelinghuys, P, Georgiadis, D, Dive, V, Acharya, K.R, Sturrock, E.D. | | Deposit date: | 2006-12-20 | | Release date: | 2007-05-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The structure of testis angiotensin-converting enzyme in complex with the C domain-specific inhibitor RXPA380.
Biochemistry, 46, 2007
|
|
