3S8L
 
 | |
3S8O
 
 | |
4DG5
 
 | |
4F95
 
 | |
7N6K
 
 | | Crystal structure of the substrate-binding domain of E. coli DnaK in complex with the peptide RALALLPLSR | | Descriptor: | Alkaline phosphatase peptide, Chaperone protein DnaK, SULFATE ION | | Authors: | Jansen, R.M, Ozden, C, Gierasch, L.M, Garman, S.C. | | Deposit date: | 2021-06-08 | | Release date: | 2021-06-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Selective promiscuity in the binding of E. coli Hsp70 to an unfolded protein.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7N6J
 
 | | Crystal structure of the substrate-binding domain of E. coli DnaK in complex with the peptide RKQSTIALALLPLLFTPRR | | Descriptor: | Alkaline phosphatase peptide, Chaperone protein DnaK, SULFATE ION | | Authors: | Jansen, R.M, Ozden, C, Gierasch, L.M, Garman, S.C. | | Deposit date: | 2021-06-08 | | Release date: | 2021-06-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Selective promiscuity in the binding of E. coli Hsp70 to an unfolded protein.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7N6L
 
 | | Crystal structure of the substrate-binding domain of E. coli DnaK in complex with the peptide EANQQKPLLGLFADG | | Descriptor: | Alkaline phosphatase peptide, Chaperone protein DnaK, GLYCEROL, ... | | Authors: | Jansen, R.M, Ozden, C, Gierasch, L.M, Garman, S.C. | | Deposit date: | 2021-06-08 | | Release date: | 2021-06-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Selective promiscuity in the binding of E. coli Hsp70 to an unfolded protein.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
3UEF
 
 | | Crystal structure of human Survivin bound to histone H3 (C2 space group). | | Descriptor: | 1,2-ETHANEDIOL, Baculoviral IAP repeat-containing protein 5, N-terminal fragment of histone H3, ... | | Authors: | Niedzialkowska, E, Porebski, P.J, Wang, F, Higgins, J.M, Stukenberg, P.T, Minor, W. | | Deposit date: | 2011-10-30 | | Release date: | 2012-03-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Molecular basis for phosphospecific recognition of histone H3 tails by Survivin paralogues at inner centromeres.
Mol.Biol.Cell, 23, 2012
|
|
5D3Z
 
 | | Crystal structure of the thioesterase domain of deoxyerythronolide B synthase in complex with a small phosphonate inhibitor | | Descriptor: | CALCIUM ION, Erythronolide synthase, modules 5 and 6, ... | | Authors: | Bergeret, F, Argyropoulos, P, Boddy, C.N, Schmeing, T.M. | | Deposit date: | 2015-08-06 | | Release date: | 2015-12-09 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Towards a characterization of the structural determinants of specificity in the macrocyclizing thioesterase for deoxyerythronolide B biosynthesis.
Biochim.Biophys.Acta, 1860, 2015
|
|
3UED
 
 | | Crystal structure of human Survivin bound to histone H3 phosphorylated on threonine-3 (C2 space group). | | Descriptor: | Baculoviral IAP repeat-containing protein 5, N-terminal fragment of histone H3, ZINC ION | | Authors: | Niedzialkowska, E, Porebski, P.J, Wang, F, Higgins, J.M, Stukenberg, P.T, Minor, W. | | Deposit date: | 2011-10-30 | | Release date: | 2012-03-07 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Molecular basis for phosphospecific recognition of histone H3 tails by Survivin paralogues at inner centromeres.
Mol.Biol.Cell, 23, 2012
|
|
3UEE
 
 | | Crystal structure of human Survivin K62A mutant bound to N-terminal histone H3 | | Descriptor: | Baculoviral IAP repeat-containing protein 5, N-terminal fragment of histone H3, ZINC ION | | Authors: | Niedzialkowska, E, Porebski, P.J, Wang, F, Higgins, J.M, Stukenberg, P.T, Minor, W. | | Deposit date: | 2011-10-30 | | Release date: | 2012-03-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Molecular basis for phosphospecific recognition of histone H3 tails by Survivin paralogues at inner centromeres.
Mol.Biol.Cell, 23, 2012
|
|
5DOG
 
 | | Crystal structure of human carbonic anhydrase isozyme II with 2-(Benzylamino)-3,5,6-trifluoro-4-[(2-phenylethyl)thio]benzene- sulfonamide | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-(benzylamino)-3,5,6-trifluoro-4-[(2-phenylethyl)sulfanyl]benzenesulfonamide, BICINE, ... | | Authors: | Smirnov, A, Manakova, E, Grazulis, S. | | Deposit date: | 2015-09-11 | | Release date: | 2016-09-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Intrinsic Thermodynamics and Structures of 2,4- and 3,4-Substituted Fluorinated Benzenesulfonamides Binding to Carbonic Anhydrases.
ChemMedChem, 12, 2017
|
|
7KQF
 
 | |
4FNI
 
 | |
5DMU
 
 | | Structure of the NHEJ polymerase from Methanocella paludicola | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, NHEJ Polymerase, ... | | Authors: | Brissett, N.C, Bartlett, E.J, Doherty, A.J. | | Deposit date: | 2015-09-09 | | Release date: | 2015-10-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.949 Å) | | Cite: | Molecular basis for DNA strand displacement by NHEJ repair polymerases.
Nucleic Acids Res., 44, 2016
|
|
3UVP
 
 | | Human p38 MAP Kinase in Complex with a Benzamide Substituted Benzosuberone | | Descriptor: | Mitogen-activated protein kinase 14, N-{2-fluoro-5-[(5-oxo-6,7,8,9-tetrahydro-5H-benzo[7]annulen-2-yl)amino]phenyl}benzamide, octyl beta-D-glucopyranoside | | Authors: | Mayer-Wrangowski, S.C, Richters, A, Gruetter, C, Rauh, D. | | Deposit date: | 2011-11-30 | | Release date: | 2012-11-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Targeting the Hinge Glycine Flip and the Activation Loop: Novel Approach to Potent p38 alpha Inhibitors.
J.Med.Chem., 55, 2012
|
|
3UVR
 
 | | Human p38 MAP Kinase in Complex with KM064 | | Descriptor: | 1-[3-tert-butyl-1-(4-methylphenyl)-1H-pyrazol-5-yl]-3-{3-[(5-oxo-6,7,8,9-tetrahydro-5H-benzo[7]annulen-2-yl)amino]phenyl}urea, Mitogen-activated protein kinase 14, octyl beta-D-glucopyranoside | | Authors: | Richters, A, Mayer-Wrangowski, S.C, Gruetter, C, Rauh, D. | | Deposit date: | 2011-11-30 | | Release date: | 2012-12-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: |
|
|
3V03
 
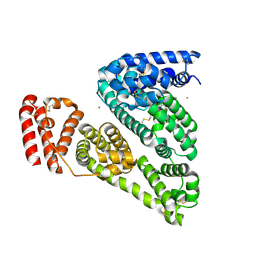 | | Crystal structure of Bovine Serum Albumin | | Descriptor: | ACETATE ION, CALCIUM ION, Serum albumin | | Authors: | Majorek, K.A, Porebski, P.J, Chruszcz, M, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2011-12-07 | | Release date: | 2012-01-04 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and immunologic characterization of bovine, horse, and rabbit serum albumins.
Mol.Immunol., 52, 2012
|
|
3UVQ
 
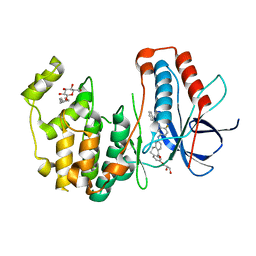 | | Human p38 MAP Kinase in Complex with a Dibenzosuberone Derivative | | Descriptor: | Mitogen-activated protein kinase 14, N-{5-[(7-{[(2R)-2,3-dihydroxypropyl]oxy}-5-oxo-10,11-dihydro-5H-dibenzo[a,d][7]annulen-2-yl)amino]-2-fluorophenyl}benzamide, octyl beta-D-glucopyranoside | | Authors: | Mayer-Wrangowski, S.C, Richters, A, Gruetter, C, Rauh, D. | | Deposit date: | 2011-11-30 | | Release date: | 2012-12-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Dibenzosuberones as p38 mitogen-activated protein kinase inhibitors with low ATP competitiveness and outstanding whole blood activity.
J.Med.Chem., 56, 2013
|
|
2JE2
 
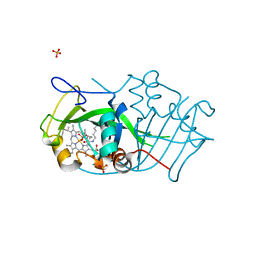 | | Cytochrome P460 from Nitrosomonas europaea - probable nonphysiological oxidized form | | Descriptor: | CYTOCHROME P460, HEME C, PHOSPHATE ION | | Authors: | Pearson, A.R, Elmore, B.O, Yang, C, Ferrara, J.D, Hooper, A.B, Wilmot, C.M. | | Deposit date: | 2007-01-13 | | Release date: | 2007-07-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Crystal Structure of Cytochrome P460 of Nitrosomonas Europaea Reveals a Novel Cytochrome Fold and Heme-Protein Cross-Link.
Biochemistry, 46, 2007
|
|
2JE3
 
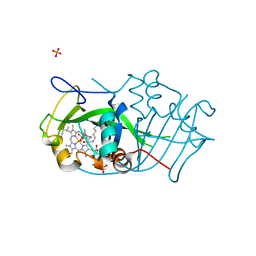 | | Cytochrome P460 from Nitrosomonas europaea - probable physiological form | | Descriptor: | CYTOCHROME P460, HEME C, PHOSPHATE ION | | Authors: | Pearson, A.R, Elmore, B.O, Yang, C, Ferrara, J.D, Hooper, A.B, Wilmot, C.M. | | Deposit date: | 2007-01-13 | | Release date: | 2007-07-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Crystal Structure of Cytochrome P460 of Nitrosomonas Europaea Reveals a Novel Cytochrome Fold and Heme-Protein Cross-Link.
Biochemistry, 46, 2007
|
|
3UEG
 
 | | Crystal structure of human Survivin K62A mutant | | Descriptor: | 1,2-ETHANEDIOL, Baculoviral IAP repeat-containing protein 5, TETRAETHYLENE GLYCOL, ... | | Authors: | Niedzialkowska, E, Porebski, P.J, Wang, F, Higgins, J.M, Stukenberg, P.T, Minor, W. | | Deposit date: | 2011-10-30 | | Release date: | 2012-03-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Molecular basis for phosphospecific recognition of histone H3 tails by Survivin paralogues at inner centromeres.
Mol.Biol.Cell, 23, 2012
|
|
3UEI
 
 | | Crystal structure of human Survivin E65A mutant | | Descriptor: | Baculoviral IAP repeat-containing protein 5, ZINC ION | | Authors: | Niedzialkowska, E, Porebski, P.J, Wang, F, Higgins, J.M, Stukenberg, P.T, Minor, W. | | Deposit date: | 2011-10-30 | | Release date: | 2012-03-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Molecular basis for phosphospecific recognition of histone H3 tails by Survivin paralogues at inner centromeres.
Mol Biol Cell, 23, 2012
|
|
3UEH
 
 | | Crystal structure of human Survivin H80A mutant | | Descriptor: | 1,2-ETHANEDIOL, Baculoviral IAP repeat-containing protein 5, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Niedzialkowska, E, Porebski, P.J, Wang, F, Higgins, J.M, Stukenberg, P.T, Minor, W. | | Deposit date: | 2011-10-30 | | Release date: | 2012-03-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Molecular basis for phosphospecific recognition of histone H3 tails by Survivin paralogues at inner centromeres.
Mol Biol Cell, 23, 2012
|
|
3V09
 
 | | Crystal structure of Rabbit Serum Albumin | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Majorek, K.A, Porebski, P.J, Chruszcz, M, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2011-12-07 | | Release date: | 2012-01-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Structural and immunologic characterization of bovine, horse, and rabbit serum albumins.
Mol.Immunol., 52, 2012
|
|
