5I9J
 
 | | Structure of the cholesterol and lutein-binding domain of human STARD3 at 1.74A | | Descriptor: | 1,2-ETHANEDIOL, L(+)-TARTARIC ACID, SULFATE ION, ... | | Authors: | Horvath, M.P, Bernstein, P.S, Li, B, George, E.W, Tran, Q.T. | | Deposit date: | 2016-02-20 | | Release date: | 2016-03-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Structure of the lutein-binding domain of human StARD3 at 1.74 angstrom resolution and model of a complex with lutein.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
5ILY
 
 | | Tobacco 5-epi-aristolochene synthase with BIS-TRIS buffer molecule and diphosphate (PPi) | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5-epi-aristolochene synthase, DIPHOSPHATE, ... | | Authors: | Koo, H.J, Louie, G.V, Xu, Y, Bowman, M, Noel, J.P. | | Deposit date: | 2016-03-05 | | Release date: | 2017-03-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Small-molecule buffer components can directly affect terpene-synthase activity by interacting with the substrate-binding site of the enzyme
To Be Published
|
|
5IM9
 
 | | Xanthomonas campestris Peroxiredoxin Q - Structure F1 | | Descriptor: | Bacterioferritin comigratory protein, SODIUM ION | | Authors: | Perkins, A, Parsonage, D, Nelson, K.J, Poole, L.B, Karplus, A. | | Deposit date: | 2016-03-05 | | Release date: | 2016-09-21 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Peroxiredoxin Catalysis at Atomic Resolution.
Structure, 24, 2016
|
|
2XZ2
 
 | |
5IAY
 
 | | NMR structure of UHRF1 Tandem Tudor Domains in a complex with Spacer peptide | | Descriptor: | E3 ubiquitin-protein ligase UHRF1, Spacer | | Authors: | Fang, J, Cheng, J, Wang, J, Zhang, Q, Liu, M, Gong, R, Wang, P, Zhang, X, Feng, Y, Lan, W, Gong, Z, Tang, C, Wong, J, Yang, H, Cao, C, Xu, Y. | | Deposit date: | 2016-02-22 | | Release date: | 2016-04-20 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Hemi-methylated DNA opens a closed conformation of UHRF1 to facilitate its histone recognition
Nat Commun, 7, 2016
|
|
5IC1
 
 | | Structural analysis of a talin triple domain module, E1794Y, E1797Y, Q1801Y mutant | | Descriptor: | 1,2-ETHANEDIOL, Talin-1 | | Authors: | Wu, J, Chang, Y.-C.E, Zhang, H, Huang, Q.-Q. | | Deposit date: | 2016-02-22 | | Release date: | 2016-05-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and Functional Analysis of a Talin Triple-Domain Module Suggests an Alternative Talin Autoinhibitory Configuration.
Structure, 24, 2016
|
|
5US3
 
 | |
5IME
 
 | | Crystal structure of P21-activated kinase 1 (PAK1) in complex with compound 9 | | Descriptor: | 8-(3-aminopropyl)-6-[2-chloro-4-(3-methyl-2-oxopyrazin-1(2H)-yl)phenyl]-2-(methylamino)pyrido[2,3-d]pyrimidin-7(8H)-one, Serine/threonine-protein kinase PAK 1 | | Authors: | Li, D, Wang, W. | | Deposit date: | 2016-03-06 | | Release date: | 2016-05-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.217 Å) | | Cite: | Chemically Diverse Group I p21-Activated Kinase (PAK) Inhibitors Impart Acute Cardiovascular Toxicity with a Narrow Therapeutic Window.
J.Med.Chem., 59, 2016
|
|
2XBD
 
 | | INTERNAL XYLAN BINDING DOMAIN FROM CELLULOMONAS FIMI XYLANASE D, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | XYLANASE D | | Authors: | Simpson, P.J, Bolam, D.N, Cooper, A, Ciruela, A, Hazlewood, G.P, Gilbert, H.J, Williamson, M.P. | | Deposit date: | 1998-10-27 | | Release date: | 1999-07-21 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | A family IIb xylan-binding domain has a similar secondary structure to a homologous family IIa cellulose-binding domain but different ligand specificity.
Structure Fold.Des., 7, 1999
|
|
5VAV
 
 | |
7M94
 
 | | Bovine sigma-2 receptor bound to Roluperidone | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Roluperidone, Sigma intracellular receptor 2 | | Authors: | Alon, A, Kruse, A.C. | | Deposit date: | 2021-03-30 | | Release date: | 2021-12-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Structures of the sigma 2 receptor enable docking for bioactive ligand discovery.
Nature, 600, 2021
|
|
5UZF
 
 | | Insights into Watson-Crick/Hoogsteen Breathing Dynamics and Damage Repair from the Solution Structure and Dynamic Ensemble of DNA Duplexes containing m1A - A6-DNA structure | | Descriptor: | DNA (5'-D(*CP*GP*AP*TP*TP*TP*TP*TP*TP*GP*GP*C)-3'), DNA (5'-D(*GP*CP*CP*AP*AP*AP*AP*AP*AP*TP*CP*G)-3') | | Authors: | Sathyamoorthy, B, Shi, H, Xue, Y, Al-Hashimi, H.M. | | Deposit date: | 2017-02-26 | | Release date: | 2017-04-05 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Insights into Watson-Crick/Hoogsteen breathing dynamics and damage repair from the solution structure and dynamic ensemble of DNA duplexes containing m1A.
Nucleic Acids Res., 45, 2017
|
|
5IMO
 
 | | Nanobody targeting mouse Vsig4 in Spacegroup P3221 | | Descriptor: | Nanobody, Protein Vsig4 | | Authors: | Wen, Y, Zheng, F. | | Deposit date: | 2016-03-06 | | Release date: | 2017-01-11 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural evaluation of a nanobody targeting complement receptor Vsig4 and its cross reactivity
Immunobiology, 222, 2017
|
|
7M96
 
 | | Bovine sigma-2 receptor bound to Z4857158944 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-({[(1S)-1-hydroxy-2-methyl-1-phenylpropan-2-yl]amino}methyl)-1-methyl-3,4-dihydroquinolin-2(1H)-one, ... | | Authors: | Alon, A, Kruse, A.C. | | Deposit date: | 2021-03-30 | | Release date: | 2021-12-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Structures of the sigma 2 receptor enable docking for bioactive ligand discovery.
Nature, 600, 2021
|
|
5IMU
 
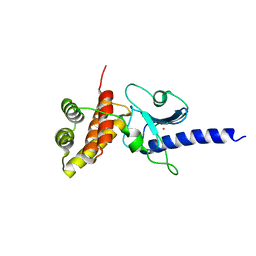 | | A fragment of conserved hypothetical protein Rv3899c (residues 184-410) from Mycobacterium tuberculosis | | Descriptor: | POTASSIUM ION, Tat (Twin-arginine translocation) pathway signal sequence containing protein | | Authors: | Li, D.F, Gao, Y.R, Liu, Y.Y, Bi, L.J. | | Deposit date: | 2016-03-07 | | Release date: | 2017-03-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of Rv3899c(184-410), a hypothetical protein from Mycobacterium tuberculosis
Acta Crystallogr F Struct Biol Commun, 72, 2016
|
|
5V1E
 
 | |
5VC7
 
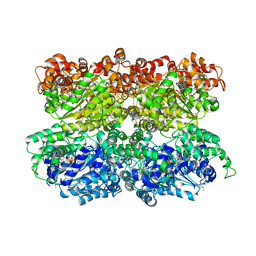 | | VCP like ATPase from T. acidophilum (VAT) - conformation 1 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, VCP-like ATPase | | Authors: | Ripstein, Z.A, Huang, R, Augustyniak, R, Kay, L.E, Rubinstein, J.L. | | Deposit date: | 2017-03-31 | | Release date: | 2017-04-26 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structure of a AAA+ unfoldase in the process of unfolding substrate.
Elife, 6, 2017
|
|
5IMZ
 
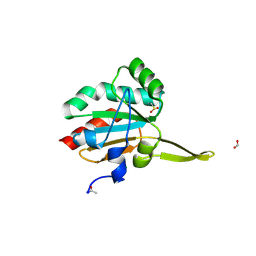 | | Xanthomonas campestris Peroxiredoxin Q - Structure F7 | | Descriptor: | Bacterioferritin comigratory protein, CHLORIDE ION, FORMIC ACID, ... | | Authors: | Perkins, A, Parsonage, D, Nelson, K.J, Poole, L.B, Karplus, A. | | Deposit date: | 2016-03-07 | | Release date: | 2016-09-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Peroxiredoxin Catalysis at Atomic Resolution.
Structure, 24, 2016
|
|
5UZT
 
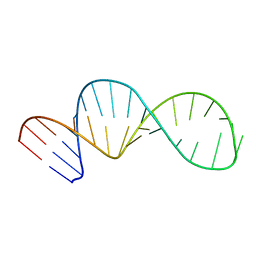 | |
7LO9
 
 | | RNA dodecamer containing a GNA A residue | | Descriptor: | Chains: A,B,C,D | | Authors: | Harp, J.M, Wawrzak, Z, Egli, M. | | Deposit date: | 2021-02-09 | | Release date: | 2021-12-22 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Overcoming GNA/RNA base-pairing limitations using isonucleotides improves the pharmacodynamic activity of ESC+ GalNAc-siRNAs.
Nucleic Acids Res., 49, 2021
|
|
5UJG
 
 | | ovGRN12-35_3s | | Descriptor: | Granulin | | Authors: | Bansal, P, Smout, M, Wilson, D, Caceres, C.C, Dastpeyman, M, Sotillo, J, Seifert, J, Brindley, P, Loukas, A, Daly, N. | | Deposit date: | 2017-01-17 | | Release date: | 2018-01-24 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Development of a Potent Wound Healing Agent Based on the Liver Fluke Granulin Structural Fold.
J. Med. Chem., 60, 2017
|
|
5IN7
 
 | | X-RAY STRUCTURE OF THE N-TERMINAL DOMAIN OF HUMAN DOUBLECORTIN | | Descriptor: | Neuronal migration protein doublecortin | | Authors: | Ruf, A, Benz, J, Burger, D, D'Arcy, B, Debulpaep, M, Di Lello, P, Fry, D, Huber, W, Kremer, T, Laeremans, T, Matile, H, Ross, A, Rudolph, M.G, Rufer, A.C, Sharma, A, Steinmetz, M.O, Steyaert, J, Schoch, G, Stihle, M, Thoma, R. | | Deposit date: | 2016-03-07 | | Release date: | 2016-03-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Crystal Structures of the Human Doublecortin C- and N-terminal Domains in Complex with Specific Antibodies.
J.Biol.Chem., 291, 2016
|
|
1SBD
 
 | | SOYBEAN AGGLUTININ COMPLEXED WITH 2,4-PENTASACCHARIDE | | Descriptor: | CALCIUM ION, MANGANESE (II) ION, SOYBEAN AGGLUTININ, ... | | Authors: | Dessen, A, Olsen, L.R, Gupta, D, Sabesan, S, Brewer, C.F, Sacchettini, J.C. | | Deposit date: | 1995-07-13 | | Release date: | 1998-04-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | X-ray crystallographic studies of unique cross-linked lattices between four isomeric biantennary oligosaccharides and soybean agglutinin.
Biochemistry, 36, 1997
|
|
5UJL
 
 | |
2Y1S
 
 | | Microvirin lectin | | Descriptor: | Mannan-binding lectin | | Authors: | Bewley, C.A, Hussan, S. | | Deposit date: | 2010-12-10 | | Release date: | 2011-04-06 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the monovalent lectin microvirin in complex with Man(alpha)(1-2)Man provides a basis for anti-HIV activity with low toxicity.
J. Biol. Chem., 286, 2011
|
|
