5V90
 
 | |
7LPX
 
 | | Crystal Structure of HIV-1 RT in Complex with NBD-14270 | | Descriptor: | Reverse transcriptase p51, Reverse transcriptase p66, SULFATE ION, ... | | Authors: | Losada, N, Ruiz, F.X, Arnold, E. | | Deposit date: | 2021-02-12 | | Release date: | 2021-11-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | HIV-1 gp120 Antagonists Also Inhibit HIV-1 Reverse Transcriptase by Bridging the NNRTI and NRTI Sites.
J.Med.Chem., 64, 2021
|
|
5IHU
 
 | | Crystal structure of bovine Fab B11 | | Descriptor: | bovine Fab B11 heavy chain, bovine Fab B11 light chain | | Authors: | Stanfield, R, Wilson, I. | | Deposit date: | 2016-02-29 | | Release date: | 2016-10-05 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Conservation and diversity in the ultralong third heavy-chain complementarity-determining region of bovine antibodies.
Sci Immunol, 1, 2016
|
|
7LQU
 
 | | Crystal Structure of HIV-1 RT in Complex with NBD-14075 | | Descriptor: | Reverse transcriptase p51, Reverse transcriptase p66, SULFATE ION, ... | | Authors: | Losada, N, Ruiz, F.X, Gruber, K, Das, K, Arnold, E. | | Deposit date: | 2021-02-15 | | Release date: | 2021-11-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | HIV-1 gp120 Antagonists Also Inhibit HIV-1 Reverse Transcriptase by Bridging the NNRTI and NRTI Sites.
J.Med.Chem., 64, 2021
|
|
5I1I
 
 | | CRYSTAL STRUCTURE OF HUMAN GERMLINE ANTIBODY IGHV3-53/IGKV4-1 | | Descriptor: | FAB HEAVY CHAIN, FAB LIGHT CHAIN | | Authors: | Teplyakov, A, Obmolova, G, Malia, T, Luo, J, Gilliland, G. | | Deposit date: | 2016-02-05 | | Release date: | 2016-06-08 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural diversity in a human antibody germline library.
Mabs, 8, 2016
|
|
5VA9
 
 | |
7LJ3
 
 | | Structure of human transfer RNA visualized in the cytomegalovirus, a DNA virus | | Descriptor: | RNA (75-MER), Tegument protein pp150 | | Authors: | Liu, Y.T, Strugatsky, D, Liu, W, Zhou, Z.H. | | Deposit date: | 2021-01-28 | | Release date: | 2021-11-17 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structure of human cytomegalovirus virion reveals host tRNA binding to capsid-associated tegument protein pp150.
Nat Commun, 12, 2021
|
|
6PPG
 
 | |
5I1U
 
 | |
2Y3G
 
 | | Se-Met form of Cupriavidus metallidurans CH34 CnrXs | | Descriptor: | CHLORIDE ION, GLYCEROL, NICKEL AND COBALT RESISTANCE PROTEIN CNRR, ... | | Authors: | Trepreau, J, Girard, E, Maillard, A.P, de Rosny, E, Petit-Haertlein, I, Kahn, R, Coves, J. | | Deposit date: | 2010-12-20 | | Release date: | 2011-03-30 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Structural Basis for Metal Sensing by Cnrx.
J.Mol.Biol., 408, 2011
|
|
5I2D
 
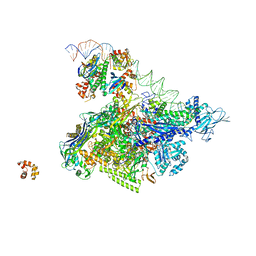 | | Crystal structure of T. thermophilus TTHB099 class II transcription activation complex: TAP-RPo | | Descriptor: | DNA (72-MER), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Feng, Y, Zhang, Y, Ebright, R.H. | | Deposit date: | 2016-02-08 | | Release date: | 2016-06-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (4.405 Å) | | Cite: | Structural basis of transcription activation.
Science, 352, 2016
|
|
5II6
 
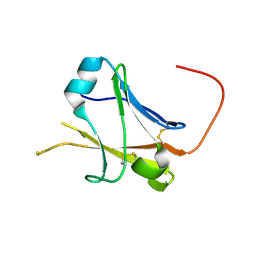 | | Crystal structure of the ZP-N1 domain of mouse sperm receptor ZP2 at 0.95 A resolution | | Descriptor: | Zona pellucida sperm-binding protein 2 | | Authors: | Dioguardi, E, Han, L, Nishimura, K, De Sanctis, D, Jovine, L. | | Deposit date: | 2016-03-01 | | Release date: | 2017-06-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Structural Basis of Egg Coat-Sperm Recognition at Fertilization.
Cell, 169, 2017
|
|
5V7B
 
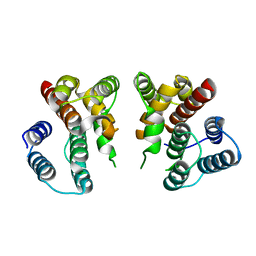 | | Crystal structure of Influenza A virus matrix protein M1 (NLS-88E) | | Descriptor: | Matrix protein 1 | | Authors: | Musayev, F.N, Safo, M.K, Desai, U.R, Xie, H, Mosier, P.D, Chiang, M.-J. | | Deposit date: | 2017-03-20 | | Release date: | 2017-04-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Maintaining pH-dependent conformational flexibility of M1 is critical for efficient influenza A virus replication.
Emerg Microbes Infect, 6, 2017
|
|
7M78
 
 | | Room Temperature XFEL Crystallography reveals asymmetry in the vicinity of the two phylloquinones in Photosystem I | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, BETA-CAROTENE, ... | | Authors: | Keable, S.M, Simon, P.S, Kolsch, A, Kern, J, Yachandra, V.K, Zouni, A, Yano, J. | | Deposit date: | 2021-03-26 | | Release date: | 2021-11-24 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Room temperature XFEL crystallography reveals asymmetry in the vicinity of the two phylloquinones in photosystem I.
Sci Rep, 11, 2021
|
|
5I58
 
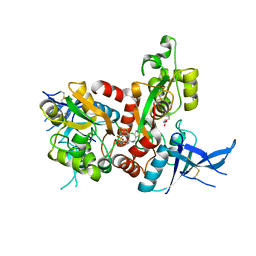 | | GLUTAMATE- AND GLYCINE-BOUND GLUN1/GLUN2A AGONIST BINDING DOMAINS WITH MPX-004 | | Descriptor: | 5-({[(3-chloro-4-fluorophenyl)sulfonyl]amino}methyl)-N-[(2-methyl-1,3-thiazol-5-yl)methyl]pyrazine-2-carboxamide, GLUTAMIC ACID, GLYCINE, ... | | Authors: | Mou, T.-C, Sprang, S.R, Hansen, K.B. | | Deposit date: | 2016-02-14 | | Release date: | 2016-09-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Structural Basis for Negative Allosteric Modulation of GluN2A-Containing NMDA Receptors.
Neuron, 91, 2016
|
|
5I5O
 
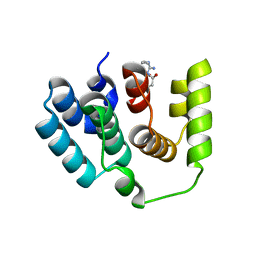 | |
5VB2
 
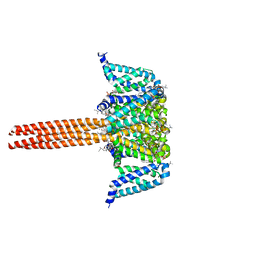 | |
5IIE
 
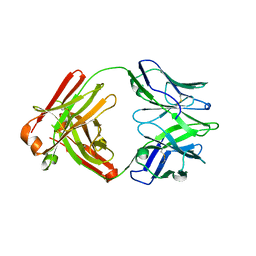 | |
5V8E
 
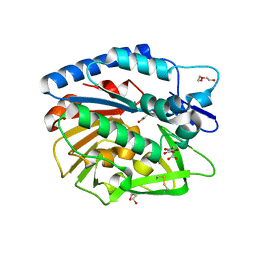 | | Structure of Bacillus cereus PatB1 | | Descriptor: | Bacillus cereus PatB1, CITRIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Sychantha, D, Little, D.J, Chapman, R.N, Boons, G.J, Robinson, H, Howell, P.L, Clarke, A.J. | | Deposit date: | 2017-03-21 | | Release date: | 2017-10-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | PatB1 is an O-acetyltransferase that decorates secondary cell wall polysaccharides.
Nat. Chem. Biol., 14, 2018
|
|
7LJI
 
 | | Structure of poly(aspartic acid) hydrolase PahZ2 with Gd+3 bound | | Descriptor: | GADOLINIUM ION, Poly(Aspartic acid) hydrolase | | Authors: | Brambley, C.A, Yared, T.J, Gonzalez, M, Jansch, A.L, Wallen, J.R, Weiland, M.H, Miller, J.M. | | Deposit date: | 2021-01-29 | | Release date: | 2021-12-08 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Sphingomonas sp. KT-1 PahZ2 Structure Reveals a Role for Conformational Dynamics in Peptide Bond Hydrolysis.
J.Phys.Chem.B, 125, 2021
|
|
5V8U
 
 | |
7M75
 
 | | Room Temperature XFEL Crystallography reveals asymmetry in the vicinity of the two phylloquinones in Photosystem I | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, BETA-CAROTENE, ... | | Authors: | Keable, S.M, Simon, P.S, Kolsch, A, Kern, J, Yachandra, V.K, Zouni, A, Yano, J. | | Deposit date: | 2021-03-26 | | Release date: | 2021-11-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Room temperature XFEL crystallography reveals asymmetry in the vicinity of the two phylloquinones in photosystem I.
Sci Rep, 11, 2021
|
|
2XOA
 
 | |
7M76
 
 | | Room Temperature XFEL Crystallography reveals asymmetry in the vicinity of the two phylloquinones in Photosystem I | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, BETA-CAROTENE, ... | | Authors: | Keable, S.M, Simon, P.S, Kolsch, A, Kern, J, Yachandra, V.K, Zouni, A, Yano, J. | | Deposit date: | 2021-03-26 | | Release date: | 2021-11-24 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Room temperature XFEL crystallography reveals asymmetry in the vicinity of the two phylloquinones in photosystem I.
Sci Rep, 11, 2021
|
|
2Y43
 
 | |
