7KRB
 
 | |
6UYJ
 
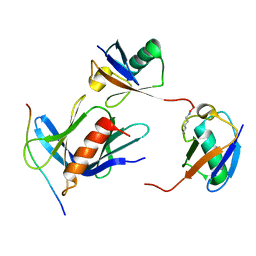 | | hRpn13:hRpn2:K48-diubiquitin | | Descriptor: | 26S proteasome non-ATPase regulatory subunit 1, Proteasomal ubiquitin receptor ADRM1, Ubiquitin | | Authors: | Lu, X, Walters, K.J. | | Deposit date: | 2019-11-13 | | Release date: | 2020-03-11 | | Last modified: | 2020-05-20 | | Method: | SOLUTION NMR | | Cite: | An Extended Conformation for K48 Ubiquitin Chains Revealed by the hRpn2:Rpn13:K48-Diubiquitin Structure.
Structure, 28, 2020
|
|
6UYI
 
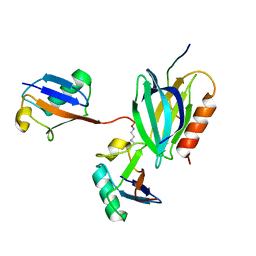 | | hRpn13:hRpn2:K48-diubiquitin | | Descriptor: | 26S proteasome non-ATPase regulatory subunit 1, Proteasomal ubiquitin receptor ADRM1, Ubiquitin | | Authors: | Lu, X, Walters, K.J. | | Deposit date: | 2019-11-13 | | Release date: | 2020-03-11 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | An Extended Conformation for K48 Ubiquitin Chains Revealed by the hRpn2:Rpn13:K48-Diubiquitin Structure.
Structure, 28, 2020
|
|
6VGT
 
 | |
8UM9
 
 | |
8UMB
 
 | |
8UM7
 
 | |
6VLJ
 
 | |
3ZKT
 
 | | SOLUTION STRUCTURE OF THE SOMATOSTATIN SST3 RECEPTOR ANTAGONIST TAU- CONOTOXIN CnVA | | Descriptor: | TAU-CNVA | | Authors: | Petrel, C, Hocking, H.G, Reynaud, M, Favreau, P, Paolini-Bertrand, M, Peigneur, S, Upert, G, Tytgat, J, Gilles, N, Hartley, O, Boelens, R, Stocklin, R, Servent, D. | | Deposit date: | 2013-01-24 | | Release date: | 2013-04-24 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Identification, Structural and Pharmacological Characterization of Tau-Cnva, a Conopeptide that Selectively Interacts with Somatostatin Sst3 Receptor.
Biochem.Pharmacol, 85, 2013
|
|
8UMS
 
 | |
8UMA
 
 | |
6VJQ
 
 | |
3ZJ2
 
 | | Structure of Nab2p tandem zinc finger 34 | | Descriptor: | NUCLEAR POLYADENYLATED RNA-BINDING PROTEIN NAB2, ZINC ION | | Authors: | Martinez-Lumbreras, S, Santiveri, C.M, Mirassou, Y, Zorrilla, S, Perez-Canadillas, J.M. | | Deposit date: | 2013-01-16 | | Release date: | 2013-09-11 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Two Singular Types of Ccch Tandem Zinc Finger in Nab2P Contribute to Polyadenosine RNA Recognition.
Structure, 21, 2013
|
|
3ZJ1
 
 | | Structure of Nab2p tandem zinc finger 12 | | Descriptor: | NUCLEAR POLYADENYLATED RNA-BINDING PROTEIN NAB2, ZINC ION | | Authors: | Martinez-Lumbreras, S, Santiveri, C.M, Mirassou, Y, Zorrilla, S, Perez-Canadillas, J.M. | | Deposit date: | 2013-01-16 | | Release date: | 2013-09-11 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Two Singular Types of Ccch Tandem Zinc Finger in Nab2P Contribute to Polyadenosine RNA Recognition.
Structure, 21, 2013
|
|
6OCV
 
 | |
7MN1
 
 | | Rules for designing protein fold switches and their implications for the folding code | | Descriptor: | Sa1 | | Authors: | He, Y, Chen, Y, Ruan, B, Choi, J, Chen, Y, Motabar, D, Solomon, T, Simmerman, R, Kauffman, T, Gallagher, T, Bryan, P, Orban, J. | | Deposit date: | 2021-04-30 | | Release date: | 2022-05-18 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Design and characterization of a protein fold switching network.
Nat Commun, 14, 2023
|
|
7PVB
 
 | |
6AHZ
 
 | | The NMR Structure of the Polysialyltranseferase Domain (PSTD) in Polysialyltransferase ST8siaIV | | Descriptor: | CMP-N-acetylneuraminate-poly-alpha-2,8-sialyltransferase | | Authors: | Liu, X.H, Lu, B, Peng, L.X, Liao, S.M, Zhou, F, Chen, D, Lu, Z.L, Zhou, G.P, Huang, R.B. | | Deposit date: | 2018-08-21 | | Release date: | 2018-10-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The Inhibition of Polysialyltranseferase ST8SiaIV Through Heparin Binding to Polysialyltransferase Domain (PSTD).
Med Chem, 15, 2019
|
|
7NF9
 
 | | N-terminal C2H2 Zn-finger domain of Clamp | | Descriptor: | Chromatin-linked adaptor for MSL proteins, isoform A, ZINC ION | | Authors: | Mariasina, S.S, Bonchuk, A.N, Tikhonova, E.A, Efimov, S.V, Maksimenko, O.G, Georgiev, P.G, Polshakov, V.I. | | Deposit date: | 2021-02-05 | | Release date: | 2021-12-29 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structural basis for interaction between CLAMP and MSL2 proteins involved in the specific recruitment of the dosage compensation complex in Drosophila.
Nucleic Acids Res., 50, 2022
|
|
5OJT
 
 | |
7POF
 
 | |
5Y08
 
 | |
6WQE
 
 | | Solution Structure of the IWP-051-bound H-NOX from Shewanella woodyi in the Fe(II)CO ligation state | | Descriptor: | 5-fluoro-2-{1-[(2-fluorophenyl)methyl]-5-(1,2-oxazol-3-yl)-1H-pyrazol-3-yl}pyrimidin-4-ol, CARBON MONOXIDE, Heme NO binding domain protein, ... | | Authors: | Chen, C.Y, Lee, W, Montfort, W.R. | | Deposit date: | 2020-04-28 | | Release date: | 2020-07-22 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the Shewanella woodyi H-NOX protein in the presence and absence of soluble guanylyl cyclase stimulator IWP-051.
Protein Sci., 30, 2021
|
|
5V16
 
 | |
6CT1
 
 | | TFE-induced NMR structure of a novel bioactive peptide (PaDBS1R7) derived from a Pyrobaculum aerophilum ribosomal protein (L39e) | | Descriptor: | PaDBS1R7 | | Authors: | Cardoso, M.H, Chan, L.Y, Candido, E.S, Torres, M.T, Oshiro, K.G.N, Silva, I.C, Goncalves, S, Buccini, D.F, Lu, T, Santos, N.C, de la Fuente-Nunez, C, Craik, D.J, Franco, O.L. | | Deposit date: | 2018-03-21 | | Release date: | 2019-09-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | An N-capping asparagine-lysine-proline (NKP) motif contributes to a hybrid flexible/stable multifunctional peptide scaffold
Chem Sci, 13, 2022
|
|
