2OF2
 
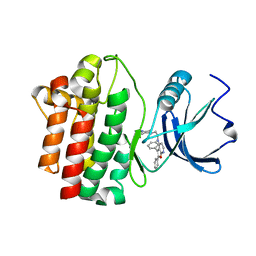 | | crystal structure of furanopyrimidine 8 bound to lck | | Descriptor: | 2,3-DIPHENYL-N-(2-PIPERAZIN-1-YLETHYL)FURO[2,3-B]PYRIDIN-4-AMINE, Proto-oncogene tyrosine-protein kinase LCK | | Authors: | Martin, M.W. | | Deposit date: | 2007-01-02 | | Release date: | 2007-02-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of novel 2,3-diarylfuro[2,3-b]pyridin-4-amines as potent and selective inhibitors of Lck: Synthesis, SAR, and pharmacokinetic properties.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
1EUY
 
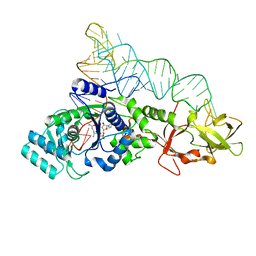 | | GLUTAMINYL-TRNA SYNTHETASE COMPLEXED WITH A TRNA MUTANT AND AN ACTIVE SITE INHIBITOR | | Descriptor: | 5'-O-[N-(L-GLUTAMINYL)-SULFAMOYL]ADENOSINE, GLUTAMINYL TRNA, GLUTAMINYL-TRNA SYNTHETASE | | Authors: | Sherlin, L.D, Bullock, T.L, Newberry, K.J, Lipman, R.S.A, Hou, Y.-M, Beijer, B, Sproat, B.S, Perona, J.J. | | Deposit date: | 2000-04-19 | | Release date: | 2000-06-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Influence of transfer RNA tertiary structure on aminoacylation efficiency by glutaminyl and cysteinyl-tRNA synthetases.
J.Mol.Biol., 299, 2000
|
|
3VHA
 
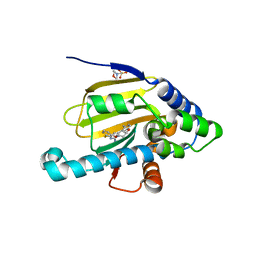 | | Hsp90 alpha N-terminal domain in complex with a macrocyclic inhibitor | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 22-methyl-13,18-dioxa-7-thia-3,5-diazatetracyclo[17.3.1.1~2,6~.1~8,12~]pentacosa-1(23),2(25),3,5,8(24),9,11,19,21-nonaen-4-amine, Heat shock protein HSP 90-alpha | | Authors: | Fukami, T.A, Ono, N. | | Deposit date: | 2011-08-24 | | Release date: | 2012-07-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Design and synthesis of novel macrocyclic 2-amino-6-arylpyrimidine Hsp90 inhibitors
Bioorg.Med.Chem.Lett., 22, 2012
|
|
6SXY
 
 | |
6SYA
 
 | |
2Y1E
 
 | | X-ray structure of 1-deoxy-D-xylulose 5-phosphate reductoisomerase, DXR, Rv2870c, from Mycobacterium tuberculosis, in complex with manganese. | | Descriptor: | 1-DEOXY-D-XYLULOSE 5-PHOSPHATE REDUCTOISOMERASE, MANGANESE (II) ION, SULFATE ION | | Authors: | Henriksson, L.M, Larsson, A.M.S, Bergfors, T, Bjorkelid, C, Unge, T, Mowbray, S.L, Jones, T.A. | | Deposit date: | 2010-12-08 | | Release date: | 2011-06-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Design, Synthesis and X-Ray Crystallographic Studies of Alpha-Aryl Substituted Fosmidomycin Analogues as Inhibitors of Mycobacterium Tuberculosis 1-Deoxy-D-Xylulose-5-Phosphate Reductoisomerase
J.Med.Chem, 54, 2011
|
|
2Y1C
 
 | | X-ray structure of 1-deoxy-D-xylulose 5-phosphate reductoisomerase, DXR, Rv2870c, from Mycobacterium tuberculosis, in complex with manganese. | | Descriptor: | 1-DEOXY-D-XYLULOSE 5-PHOSPHATE REDUCTOISOMERASE, MANGANESE (II) ION | | Authors: | Henriksson, L.M, Larsson, A.M.S, Bergfors, T, Bjorkelid, C, Unge, T, Mowbray, S.L, Jones, T.A. | | Deposit date: | 2010-12-08 | | Release date: | 2011-06-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Design, Synthesis and X-Ray Crystallographic Studies of Alpha-Aryl Substituted Fosmidomycin Analogues as Inhibitors of Mycobacterium Tuberculosis 1-Deoxy-D-Xylulose-5-Phosphate Reductoisomerase
J.Med.Chem, 54, 2011
|
|
2Y5G
 
 | | FACTOR XA - CATION INHIBITOR COMPLEX | | Descriptor: | 3-[(3AS,4R,5S,8AS,8BR)-4-[5-(5-CHLOROTHIOPHEN-2-YL)-1,3-OXAZOL-2-YL]-1,3-DIOXO-4,6,7,8,8A,8B-HEXAHYDRO-3AH-PYRROLO[3,4-A]PYRROLIZIN-2-YL]PROPYL-TRIMETHYL-AZANIUM, ACTIVATED FACTOR XA HEAVY CHAIN, FACTOR X LIGHT CHAIN, ... | | Authors: | Banner, D.W, Salonen, L.M, Holland, M.C, Haap, W, Benz, J, Diederich, F. | | Deposit date: | 2011-01-13 | | Release date: | 2011-12-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Molecular Recognition at the Active Site of Factor Xa: Cation-Pi Interactions, Stacking on Planar Peptide Surfaces, and Replacement of Structural Water.
Chemistry, 18, 2012
|
|
1EUQ
 
 | | CRYSTAL STRUCTURE OF GLUTAMINYL-TRNA SYNTHETASE COMPLEXED WITH A TRNA-GLN MUTANT AND AN ACTIVE-SITE INHIBITOR | | Descriptor: | 5'-O-[N-(L-GLUTAMINYL)-SULFAMOYL]ADENOSINE, GLUTAMINYL TRNA, GLUTAMINYL-TRNA SYNTHETASE | | Authors: | Sherlin, L.D, Bullock, T.L, Newberry, K.J, Lipman, R.S.A, Hou, Y.-M, Beijer, B, Sproat, B.S, Perona, J.J. | | Deposit date: | 2000-04-17 | | Release date: | 2000-06-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Influence of transfer RNA tertiary structure on aminoacylation efficiency by glutaminyl and cysteinyl-tRNA synthetases.
J.Mol.Biol., 299, 2000
|
|
2CKY
 
 | | Structure of the Arabidopsis thaliana thiamine pyrophosphate riboswitch with its regulatory ligand | | Descriptor: | MAGNESIUM ION, NUCLEIC ACID, OSMIUM ION, ... | | Authors: | Thore, S, Leibundgut, M, Ban, N. | | Deposit date: | 2006-04-24 | | Release date: | 2006-05-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of the Eukaryotic Thiamine Pyrophosphate Riboswitch with its Regulatory Ligand.
Science, 312, 2006
|
|
2RFS
 
 | | X-ray structure of SU11274 bound to c-Met | | Descriptor: | Hepatocyte growth factor receptor, N-(3-chlorophenyl)-N-methyl-2-oxo-3-[(3,4,5-trimethyl-1H-pyrrol-2-yl)methyl]-2H-indole-5-sulfonamide | | Authors: | Bellon, S.F, Kaplan-Lefko, P, Yang, Y, Zhang, Y, Moriguchi, J, Dussault, I. | | Deposit date: | 2007-10-01 | | Release date: | 2007-11-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | c-Met inhibitors with novel binding mode show activity against several hereditary papillary renal cell carcinoma-related mutations.
J.Biol.Chem., 283, 2008
|
|
1NC6
 
 | | Potent, small molecule inhibitors of human mast cell tryptase. Anti-asthmatic action of a dipeptide-based transition state analogue containing benzothiazole ketone | | Descriptor: | (2S,4R)-1-ACETYL-N-[(1S)-4-[(AMINOIMINOMETHYL)AMINO]-1-(2-BENZOTHIAZOLYLCARBONYL)BUTYL]-4-HYDROXY-2-PYRROLIDINECARBOXAMIDE, CALCIUM ION, SULFATE ION, ... | | Authors: | Costanzo, M.J, Yabut, S.C, Almond Jr, H.R, Andrade-Gordon, P, Corcoran, T.W, de Garavilla, L, Kauffman, J.A, Abraham, W.M, Recacha, R, Chattopadhyay, D, Maryanoff, B.E. | | Deposit date: | 2002-12-04 | | Release date: | 2003-09-23 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Potent, Small-Molecule Inhibitors of Human Mast Cell Tryptase. Antiasthmatic Action of a Dipeptide-Based Transition-State Analogue Containing a Benzothiazole Ketone.
J.Med.Chem., 46, 2003
|
|
4N6E
 
 | | Crystal structure of Amycolatopsis orientalis BexX/CysO complex | | Descriptor: | Putative thiosugar synthase, SULFATE ION, ThiS/MoaD family protein | | Authors: | Zhang, X, Zhang, Y, Kinsland, C, Sasaki, E, Sun, H.G, Lu, M.J, Liu, T, Ou, A, Li, J, Chen, Y, Liu, H, Ealick, S.E. | | Deposit date: | 2013-10-11 | | Release date: | 2014-05-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Co-opting sulphur-carrier proteins from primary metabolic pathways for 2-thiosugar biosynthesis.
Nature, 509, 2014
|
|
3VHC
 
 | | Hsp90 alpha N-terminal domain in complex with a macrocyclic inhibitor | | Descriptor: | 4-amino-20,22-dimethyl-13-oxa-7-thia-3,5,17-triazatetracyclo[17.3.1.1~2,6~.1~8,12~]pentacosa-1(23),2(25),3,5,8(24),9,11,19,21-nonaen-18-one, Heat shock protein HSP 90-alpha, MAGNESIUM ION | | Authors: | Fukami, T.A, Ono, N. | | Deposit date: | 2011-08-24 | | Release date: | 2012-07-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Design and synthesis of novel macrocyclic 2-amino-6-arylpyrimidine Hsp90 inhibitors
Bioorg.Med.Chem.Lett., 22, 2012
|
|
1JFK
 
 | | MINIMUM ENERGY REPRESENTATIVE STRUCTURE OF A CALCIUM BOUND EF-HAND PROTEIN FROM ENTAMOEBA HISTOLYTICA | | Descriptor: | CALCIUM ION, CALCIUM-BINDING PROTEIN | | Authors: | Atreya, H.S, Sahu, S.C, Bhattacharya, A, Chary, K.V.R, Govil, G. | | Deposit date: | 2001-06-21 | | Release date: | 2001-12-19 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | NMR derived solution structure of an EF-hand calcium-binding protein from Entamoeba Histolytica.
Biochemistry, 40, 2001
|
|
3E92
 
 | | Crystal Structure of P38 Kinase in Complex with A Biaryl Amide Inhibitor | | Descriptor: | GLYCEROL, Mitogen-activated protein kinase 14, N-cyclopropyl-2',6-dimethyl-4'-(5-methyl-1,3,4-oxadiazol-2-yl)biphenyl-3-carboxamide | | Authors: | Somers, D.O, Patel, S. | | Deposit date: | 2008-08-21 | | Release date: | 2008-09-30 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Kinase array design, back to front: Biaryl amides
Bioorg.Med.Chem.Lett., 18, 2008
|
|
3E93
 
 | |
393D
 
 | |
3AFQ
 
 | | Crystal structure of the single-stranded DNA binding protein from Mycobacterium leprae (Form II) | | Descriptor: | Single-stranded DNA-binding protein | | Authors: | Kaushal, P.S, Singh, P, Sharma, A, Muniyappa, K, Vijayan, M. | | Deposit date: | 2010-03-10 | | Release date: | 2010-10-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | X-ray and molecular-dynamics studies on Mycobacterium leprae single-stranded DNA-binding protein and comparison with other eubacterial SSB structures
Acta Crystallogr.,Sect.D, 66, 2010
|
|
3AFP
 
 | | Crystal structure of the single-stranded DNA binding protein from Mycobacterium leprae (Form I) | | Descriptor: | CADMIUM ION, GLYCEROL, Single-stranded DNA-binding protein | | Authors: | Kaushal, P.S, Singh, P, Sharma, A, Muniyappa, K, Vijayan, M. | | Deposit date: | 2010-03-10 | | Release date: | 2010-10-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | X-ray and molecular-dynamics studies on Mycobacterium leprae single-stranded DNA-binding protein and comparison with other eubacterial SSB structures
Acta Crystallogr.,Sect.D, 66, 2010
|
|
394D
 
 | |
1OFN
 
 | | Purification, crystallisation and preliminary structural studies of dTDP-4-keto-6-deoxy-glucose-5-epimerase (EvaD) from Amycolatopsis orientalis; the fourth enzyme in the dTDP-L-epivancosamine biosynthetic pathway. | | Descriptor: | GLYCEROL, PCZA361.16 | | Authors: | Merkel, A.B, Naismith, J.H. | | Deposit date: | 2003-04-17 | | Release date: | 2004-04-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Purification, Crystallization and Preliminary Structural Studies of Dtdp-4-Keto-6-Deoxy-Glucose-5-Epimerase (Evad) from Amycolatopsis Orientalis, the Fourth Enzyme in the Dtdp-L-Epivancosamine Biosynthetic Pathway.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
3IK2
 
 | | Crystal Structure of a Glycoside Hydrolase Family 44 Endoglucanase produced by Clostridium acetobutylium ATCC 824 | | Descriptor: | ACETATE ION, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Warner, C.D, Hoy, J.A, Ford, C.F, Honzatko, R.B, Reilly, P.J. | | Deposit date: | 2009-08-05 | | Release date: | 2009-08-18 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Tertiary structure and characterization of a glycoside hydrolase family 44 endoglucanase from Clostridium acetobutylicum.
Appl.Environ.Microbiol., 76, 2010
|
|
2RFN
 
 | | x-ray structure of c-Met with inhibitor. | | Descriptor: | 2-benzyl-5-(3-fluoro-4-{[6-methoxy-7-(3-morpholin-4-ylpropoxy)quinolin-4-yl]oxy}phenyl)-3-methylpyrimidin-4(3H)-one, Hepatocyte growth factor receptor | | Authors: | Bellon, S.F, Kaplan-Lefko, P, Yang, Y, Zhang, Y, Moriguchi, J, Dussault, I. | | Deposit date: | 2007-10-01 | | Release date: | 2007-11-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | c-Met inhibitors with novel binding mode show activity against several hereditary papillary renal cell carcinoma-related mutations.
J.Biol.Chem., 283, 2008
|
|
1JFJ
 
 | | NMR SOLUTION STRUCTURE OF AN EF-HAND CALCIUM BINDING PROTEIN FROM ENTAMOEBA HISTOLYTICA | | Descriptor: | CALCIUM-BINDING PROTEIN | | Authors: | Atreya, H.S, Sahu, S.C, Bhattacharya, A, Chary, K.V.R, Govil, G. | | Deposit date: | 2001-06-20 | | Release date: | 2001-12-19 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | NMR derived solution structure of an EF-hand calcium-binding protein from Entamoeba Histolytica.
Biochemistry, 40, 2001
|
|
