4WC3
 
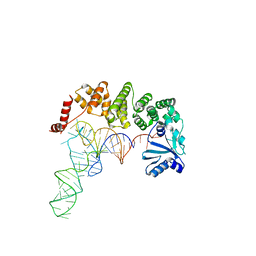 | | Structure of tRNA-processing enzyme complex 1 | | Descriptor: | Poly A polymerase, RNA 76-mer | | Authors: | Yamashita, S, Tomita, K. | | Deposit date: | 2014-09-04 | | Release date: | 2015-04-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Measurement of Acceptor-T Psi C Helix Length of tRNA for Terminal A76-Addition by A-Adding Enzyme.
Structure, 23, 2015
|
|
5HNW
 
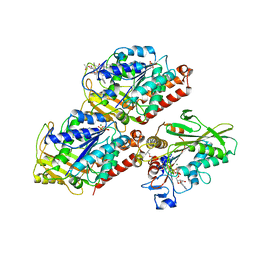 | | Structural basis of backwards motion in kinesin-14: minus-end directed nKn664 in the AMPPNP state | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Shigematsu, H, Yokoyama, T, Kikkawa, M, Shirouzu, M, Nitta, R. | | Deposit date: | 2016-01-19 | | Release date: | 2016-08-10 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (6.6 Å) | | Cite: | Structural Basis of Backwards Motion in Kinesin-1-Kinesin-14 Chimera: Implication for Kinesin-14 Motility
Structure, 24, 2016
|
|
4WC2
 
 | |
4X0A
 
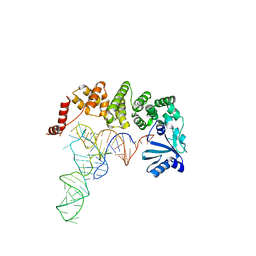 | | Structure of tRNA-processing enzyme complex 6 | | Descriptor: | Poly A polymerase, RNA (73-MER) | | Authors: | Yamashita, S, Tomita, K. | | Deposit date: | 2014-11-21 | | Release date: | 2015-04-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.505 Å) | | Cite: | Measurement of Acceptor-T Psi C Helix Length of tRNA for Terminal A76-Addition by A-Adding Enzyme.
Structure, 23, 2015
|
|
5HU3
 
 | |
4WC7
 
 | | Structure of tRNA-processing enzyme complex 5 | | Descriptor: | CYTIDINE-5'-TRIPHOSPHATE, Poly A polymerase, RNA (75-MER) | | Authors: | Yamashita, S, Tomita, K. | | Deposit date: | 2014-09-04 | | Release date: | 2015-04-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.102 Å) | | Cite: | Measurement of Acceptor-T Psi C Helix Length of tRNA for Terminal A76-Addition by A-Adding Enzyme.
Structure, 23, 2015
|
|
4UUS
 
 | | CRYSTAL STRUCTURE OF A UBX-EXD-DNA COMPLEX INCLUDING THE UBDA MOTIF | | Descriptor: | 5'-D(*AP*CP*GP*TP*GP*AP*TP*TP*TP*AP*TP*GP*GP*CP*G)-3', 5'-D(*GP*TP*CP*GP*CP*CP*AP*TP*AP*AP*AP*TP*CP*AP*C)-3', HOMEOTIC PROTEIN EXTRADENTICLE, ... | | Authors: | Foos, N, Mate, M.J, Ortiz-Lombardia, M. | | Deposit date: | 2014-07-31 | | Release date: | 2015-02-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | A Flexible Extension of the Drosophila Ultrabithorax Homeodomain Defines a Novel Hox/Pbc Interaction Mode.
Structure, 23, 2015
|
|
4UUT
 
 | |
5I7Z
 
 | | Crystal structure of a Par-6 PDZ-Crumbs 3 C-terminal peptide complex | | Descriptor: | Crb-3, DI(HYDROXYETHYL)ETHER, LD29223p | | Authors: | Whitney, D.S, Peterson, F.C, Prehoda, K.E, Volkman, B.F. | | Deposit date: | 2016-02-18 | | Release date: | 2016-03-16 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Binding of Crumbs to the Par-6 CRIB-PDZ Module Is Regulated by Cdc42.
Biochemistry, 55, 2016
|
|
4WVR
 
 | | Crystal structure of Dscam1 Ig7 domain, isoform 5 | | Descriptor: | Down syndrome cell adhesion molecule, isoform AK | | Authors: | Chen, Q, Yu, Y, Li, S, Cheng, L. | | Deposit date: | 2014-11-07 | | Release date: | 2015-11-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.948 Å) | | Cite: | Structural basis of Dscam1 homodimerization: Insights into context constraint for protein recognition
Sci Adv, 2, 2016
|
|
4WPC
 
 | |
5JSD
 
 | | Crystal structure of phiAB6 tailspike in complex with five-repeated oligosaccharides of Acinetobacter baumannii surface polysaccharide | | Descriptor: | ACETIC ACID, MALONIC ACID, beta-D-galactopyranose-(1-3)-2-amino-2-deoxy-beta-D-galactopyranose-(1-3)-[5,7-bisacetamido-3,5,7,9-tetradeoxy-L-glycero-alpha-L-manno-non-2-ulopyranosonic acid-(2-6)-beta-D-glucopyranose-(1-6)]beta-D-galactopyranose-(1-3)-2-amino-2-deoxy-beta-D-galactopyranose-(1-3)-[beta-D-glucopyranose-(1-6)]beta-D-galactopyranose, ... | | Authors: | Lee, I.M, Tu, I.F, Huang, K.F, Wu, S.H. | | Deposit date: | 2016-05-08 | | Release date: | 2017-03-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Structural basis for fragmenting the exopolysaccharide of Acinetobacter baumannii by bacteriophage Phi AB6 tailspike protein
Sci Rep, 7, 2017
|
|
2P9H
 
 | |
1B5X
 
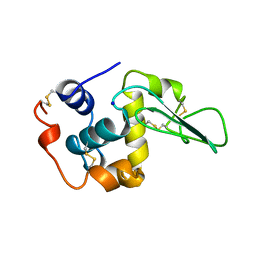 | | Contribution of hydrogen bonds to the conformational stability of human lysozyme: calorimetry and x-ray analysis of six ser->ala mutants | | Descriptor: | PROTEIN (LYSOZYME), SODIUM ION | | Authors: | Takano, K, Yamagata, Y, Kubota, M, Funahashi, J, Fujii, S, Yutani, K. | | Deposit date: | 1999-01-11 | | Release date: | 1999-01-20 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Contribution of hydrogen bonds to the conformational stability of human lysozyme: calorimetry and X-ray analysis of six Ser --> Ala mutants.
Biochemistry, 38, 1999
|
|
1B5V
 
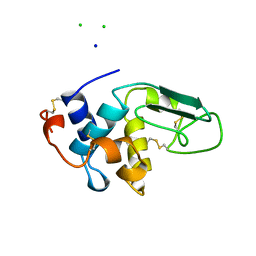 | | CONTRIBUTION OF HYDROGEN BONDS TO THE CONFORMATIONAL STABILITY OF HUMAN LYSOZYME: CALORIMETRY AND X-RAY ANALYSIS OF SIX SER->ALA MUTANTS | | Descriptor: | CHLORIDE ION, PROTEIN (LYSOZYME), SODIUM ION | | Authors: | Takano, K, Yamagata, Y, Kubota, M, Funahashi, J, Fujii, S, Yutani, K. | | Deposit date: | 1999-01-11 | | Release date: | 1999-01-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Contribution of hydrogen bonds to the conformational stability of human lysozyme: calorimetry and X-ray analysis of six Ser --> Ala mutants.
Biochemistry, 38, 1999
|
|
1B5Y
 
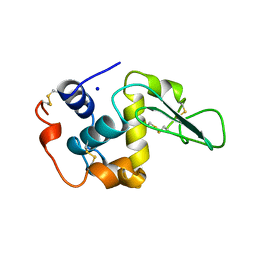 | | CONTRIBUTION OF HYDROGEN BONDS TO THE CONFORMATIONAL STABILITY OF HUMAN LYSOZYME: CALORIMETRY AND X-RAY ANALYSIS OF SIX SER->ALA MUTANTS | | Descriptor: | PROTEIN (LYSOZYME), SODIUM ION | | Authors: | Takano, K, Yamagata, Y, Kubota, M, Funahashi, J, Fujii, S, Yutani, K. | | Deposit date: | 1999-01-11 | | Release date: | 1999-01-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Contribution of hydrogen bonds to the conformational stability of human lysozyme: calorimetry and X-ray analysis of six Ser --> Ala mutants.
Biochemistry, 38, 1999
|
|
5JSE
 
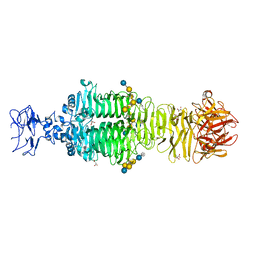 | | Crystal structure of phiAB6 tailspike in complex with three-repeated oligosaccharides of Acinetobacter baumannii surface polysaccharide | | Descriptor: | ACETIC ACID, MALONIC ACID, beta-D-galactopyranose-(1-3)-2-amino-2-deoxy-beta-D-galactopyranose-(1-3)-[5,7-bisacetamido-3,5,7,9-tetradeoxy-L-glycero-alpha-L-manno-non-2-ulopyranosonic acid-(2-6)-beta-D-glucopyranose-(1-6)]beta-D-galactopyranose-(1-3)-2-amino-2-deoxy-beta-D-galactopyranose-(1-3)-[beta-D-glucopyranose-(1-6)]beta-D-galactopyranose, ... | | Authors: | Lee, I.M, Tu, I.F, Huang, K.F, Wu, S.H. | | Deposit date: | 2016-05-08 | | Release date: | 2017-03-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structural basis for fragmenting the exopolysaccharide of Acinetobacter baumannii by bacteriophage Phi AB6 tailspike protein
Sci Rep, 7, 2017
|
|
1B5U
 
 | | CONTRIBUTION OF HYDROGEN BONDS TO THE CONFORMATIONAL STABILITY OF HUMAN LYSOZYME: CALORIMETRY AND X-RAY ANALYSIS OF SIX SER->ALA MUTANT | | Descriptor: | PROTEIN (LYSOZYME), SODIUM ION | | Authors: | Takano, K, Yamagata, Y, Kubota, M, Funahashi, J, Fujii, S, Yutani, K. | | Deposit date: | 1999-01-11 | | Release date: | 1999-01-20 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Contribution of hydrogen bonds to the conformational stability of human lysozyme: calorimetry and X-ray analysis of six Ser --> Ala mutants.
Biochemistry, 38, 1999
|
|
1B5W
 
 | | CONTRIBUTION OF HYDROGEN BONDS TO THE CONFORMATIONAL STABILITY OF HUMAN LYSOZYME: CALORIMETRY AND X-RAY ANALYSIS OF SIX SER->ALA MUTANTS | | Descriptor: | CHLORIDE ION, PROTEIN (LYSOZYME), SODIUM ION | | Authors: | Takano, K, Yamagata, Y, Kubota, M, Funahashi, J, Fujii, S, Yutani, K. | | Deposit date: | 1999-01-11 | | Release date: | 1999-01-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Contribution of hydrogen bonds to the conformational stability of human lysozyme: calorimetry and X-ray analysis of six Ser --> Ala mutants.
Biochemistry, 38, 1999
|
|
1BVO
 
 | | DORSAL HOMOLOGUE GAMBIF1 BOUND TO DNA | | Descriptor: | DNA DUPLEX, TRANSCRIPTION FACTOR GAMBIF1 | | Authors: | Cramer, P, Varrot, A, Barillas-Mury, C, Kafatos, F.C, Mueller, C.W. | | Deposit date: | 1998-09-16 | | Release date: | 1999-07-12 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of the specificity domain of the Dorsal homologue Gambif1 bound to DNA.
Structure Fold.Des., 7, 1999
|
|
2RDK
 
 | | Five site mutated Cyanovirin-N with Mannose dimer bound | | Descriptor: | Cyanovirin-N, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose | | Authors: | Fromme, R, Katiliene, Z, Fromme, P, Ghirlanda, G. | | Deposit date: | 2007-09-24 | | Release date: | 2008-05-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Conformational gating of dimannose binding to the antiviral protein cyanovirin revealed from the crystal structure at 1.35 A resolution.
Protein Sci., 17, 2008
|
|
6QF7
 
 | | Crystal structures of the recombinant beta-Factor XIIa protease with bound Thr-Arg and Pro-Arg substrate mimetics | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Coagulation factor XII, ... | | Authors: | Pathak, M, Mannal, R, Li, C, Bubacarr, G.K, Badraldin, K.H, Belviso, B.D, Camila, R.B, Dreveny, I, Fischer, P.M, Dekker, L.V, Oliva, M.L.V, Emsley, J. | | Deposit date: | 2019-01-09 | | Release date: | 2019-06-05 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Crystal structures of the recombinant beta-factor XIIa protease with bound Thr-Arg and Pro-Arg substrate mimetics.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
4U77
 
 | | BTB domain from Drosophila CP190 | | Descriptor: | Centrosome-associated zinc finger protein CP190, DIMETHYL SULFOXIDE, GLYCEROL | | Authors: | Allemand, F, Cohen-Gonsaud, M, Labesse, G, Nollmann, M. | | Deposit date: | 2014-07-30 | | Release date: | 2014-08-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Chromatin Insulator Factors Involved in Long-Range DNA Interactions and Their Role in the Folding of the Drosophila Genome.
Plos Genet., 10, 2014
|
|
6Q9M
 
 | | Central Fibronectin-III array of RIM-binding protein | | Descriptor: | PHOSPHATE ION, RIM-binding protein, isoform F | | Authors: | Driller, J.D, Habibi, S, Wahl, M.C, Loll, B. | | Deposit date: | 2018-12-18 | | Release date: | 2020-01-15 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.453 Å) | | Cite: | RIM-binding protein couples synaptic vesicle recruitment to release sites.
J.Cell Biol., 219, 2020
|
|
2SPC
 
 | | CRYSTAL STRUCTURE OF THE REPETITIVE SEGMENTS OF SPECTRIN | | Descriptor: | SPECTRIN | | Authors: | Yan, Y, Winograd, E, Viel, A, Cronin, T, Harrison, S.C, Branton, D. | | Deposit date: | 1994-03-01 | | Release date: | 1994-05-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the repetitive segments of spectrin.
Science, 262, 1993
|
|
