4CJA
 
 | | BurrH DNA-binding protein from Burkholderia rhizoxinica in complex with its target DNA | | Descriptor: | 5'-D(*DTP*AP*TP*AP*AP*CP*GP*TP*AP*TP*TP*TP*GP*CP *TP*TP*CP*TP*CP*TP*TP*AP*AP)-3', 5'-D(*DTP*TP*AP*AP*GP*AP*GP*AP*AP*GP*CP*AP*AP*DP *TP*AP*CP*GP*TP*TP*AP*TP*AP)-3', BURRH | | Authors: | Stella, S, Molina, R, Lopez-Mendez, B, Campos-Olivas, R, Duchateau, P, Montoya, G. | | Deposit date: | 2013-12-19 | | Release date: | 2014-07-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.651 Å) | | Cite: | Bud, a Helix-Loop-Helix DNA-Binding Domain for Genome Modification
Acta Crystallogr.,Sect.D, 70, 2014
|
|
5T8H
 
 | |
6HK4
 
 | | Crystal structure of GSK-3B in complex with pyrazine inhibitor C22 | | Descriptor: | 3-azanyl-6-(4-morpholin-4-ylsulfonylphenyl)-~{N}-pyridin-3-yl-pyrazine-2-carboxamide, DIMETHYL SULFOXIDE, GLY-SER-HIS-GLY-HIS-HIS-HIS-HIS-HIS, ... | | Authors: | Piretti, V, Giabbai, B, Demitri, N, Di Martino, R, Tripathi, S.K, Gobbo, D, Decherchi, S, Storici, P, Girotto, S, Cavalli, A. | | Deposit date: | 2018-09-05 | | Release date: | 2019-07-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Investigating Drug-Target Residence Time in Kinases through Enhanced Sampling Simulations.
J Chem Theory Comput, 15, 2019
|
|
6I3H
 
 | | Crystal structure of influenza A virus M1 N-terminal domain (G18A mutation) | | Descriptor: | Matrix protein 1, PHOSPHATE ION | | Authors: | Miyake, Y, Keusch, J.J, Decamps, L, Ho-Xuan, H, Iketani, S, Gut, H, Kutay, U, Helenius, A, Yamauchi, Y. | | Deposit date: | 2018-11-06 | | Release date: | 2019-09-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Influenza virus uses transportin 1 for vRNP debundling during cell entry.
Nat Microbiol, 4, 2019
|
|
8V7G
 
 | | Human DNA polymerase eta-DNA-gemC-ended primer-dAMPNPP ternary complex with Mg2+ | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]adenosine, DNA (5'-D(*AP*GP*CP*GP*TP*CP*AP*(U7B))-3'), DNA (5'-D(*CP*AP*TP*TP*GP*TP*GP*AP*CP*GP*CP*T)-3'), ... | | Authors: | Chang, C, Gao, Y. | | Deposit date: | 2023-12-04 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Human DNA polymerase eta-DNA-araC-ended primer-dAMPNPP ternary complex with Mn2+
To Be Published
|
|
6CXN
 
 | | HRFLRH peptide NMR structure | | Descriptor: | Hexapeptide HRFLRH | | Authors: | Pires, D.A.T, Arake, L.M.R, Silva, L.P, Lopez-Castillo, A, Prates, M.V, Nascimento, C.J, Bloch Jr, C. | | Deposit date: | 2018-04-03 | | Release date: | 2018-07-04 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | A previously undescribed hexapeptide His-Arg-Phe-Leu-Arg-His-NH2from amphibian skin secretion shows CO2and metal biding affinities.
Peptides, 106, 2018
|
|
6CXR
 
 | | HRFLRH peptide NMR structure in the presence of Zn(II) | | Descriptor: | Hexapeptide HRFLRH | | Authors: | Pires, D.A.T, Arake, L.M.R, Silva, L.P, Lopez-Castillo, A, Prates, M.V, Nascimento, C.J, Bloch Jr, C. | | Deposit date: | 2018-04-03 | | Release date: | 2018-07-04 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | A previously undescribed hexapeptide His-Arg-Phe-Leu-Arg-His-NH2from amphibian skin secretion shows CO2and metal biding affinities.
Peptides, 106, 2018
|
|
6CXP
 
 | | HRFLRH peptide NMR structure in the presence of Cd(II) | | Descriptor: | Hexapeptide HRFLRH | | Authors: | Pires, D.A.T, Arake, L.M.R, Silva, L.P, Lopez-Castillo, A, Prates, M.V, Nascimento, C.J, Bloch Jr, C. | | Deposit date: | 2018-04-03 | | Release date: | 2018-07-04 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | A previously undescribed hexapeptide His-Arg-Phe-Leu-Arg-His-NH2from amphibian skin secretion shows CO2and metal biding affinities.
Peptides, 106, 2018
|
|
5O1Z
 
 | |
6N62
 
 | | Escherichia coli RNA polymerase sigma70-holoenzyme bound to upstream fork promoter DNA | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Braffman, N, Hauver, J, Campbell, E.A, Darst, S.A. | | Deposit date: | 2018-11-24 | | Release date: | 2019-01-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.803 Å) | | Cite: | Structural mechanism of transcription inhibition by lasso peptides microcin J25 and capistruin.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
6J61
 
 | | Crystal Structure of Thymidylate Synthase, Thy1, from Thermus thermophilus having an Extra C Terminal Domain | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Flavin-dependent thymidylate synthase, PHOSPHATE ION | | Authors: | Ogawa, A, Sampei, G, Kawai, G. | | Deposit date: | 2019-01-12 | | Release date: | 2019-06-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the flavin-dependent thymidylate synthase Thy1 from Thermus thermophilus with an extra C-terminal domain.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
6N61
 
 | | Escherichia coli RNA polymerase sigma70-holoenzyme bound to upstream fork promoter DNA and Capistruin | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Capistruin, ... | | Authors: | Braffman, N, Hauver, J, Campbell, E.A, Darst, S.A. | | Deposit date: | 2018-11-24 | | Release date: | 2019-01-09 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.253 Å) | | Cite: | Structural mechanism of transcription inhibition by lasso peptides microcin J25 and capistruin.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
6GXF
 
 | | The hit-and-return system enables efficient time-resolved serial synchrotron crystallography: RADDAM1 | | Descriptor: | Fluoroacetate dehalogenase | | Authors: | Schulz, E.C, Mehrabi, P, Mueller-Werkmeister, H, Tellkamp, F, Stuart, W, Persch, E, De Gasparo, R, Diederich, F, Pai, E.F, Miller, R.J.D. | | Deposit date: | 2018-06-27 | | Release date: | 2018-10-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The hit-and-return system enables efficient time-resolved serial synchrotron crystallography.
Nat. Methods, 15, 2018
|
|
6CFL
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with lysyl-CAM and bound to protein Y (YfiA) at 2.6A resolution | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 16S Ribosomal RNA, 23S Ribosomal RNA, ... | | Authors: | Tereshchenkov, A.G, Dobosz-Bartoszek, M, Osterman, I.A, Marks, J, Sergeeva, V.A, Kasatsky, P, Komarova, E.S, Stavrianidi, A.N, Rodin, I.A, Konevega, A.L, Sergiev, P.V, Sumbatyan, N.V, Mankin, A.S, Bogdanov, A.A, Polikanov, Y.S. | | Deposit date: | 2018-02-15 | | Release date: | 2018-03-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Binding and Action of Amino Acid Analogs of Chloramphenicol upon the Bacterial Ribosome.
J. Mol. Biol., 430, 2018
|
|
6CFJ
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with histidyl-CAM and bound to mRNA and A-, P-, and E-site tRNAs at 2.8A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Tereshchenkov, A.G, Dobosz-Bartoszek, M, Osterman, I.A, Marks, J, Sergeeva, V.A, Kasatsky, P, Komarova, E.S, Stavrianidi, A.N, Rodin, I.A, Konevega, A.L, Sergiev, P.V, Sumbatyan, N.V, Mankin, A.S, Bogdanov, A.A, Polikanov, Y.S. | | Deposit date: | 2018-02-15 | | Release date: | 2018-03-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Binding and Action of Amino Acid Analogs of Chloramphenicol upon the Bacterial Ribosome.
J. Mol. Biol., 430, 2018
|
|
5O1W
 
 | |
5O20
 
 | | Structure of Nrd1 RNA binding domain in complex with RNA (UUAGUAAUCC) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Protein NRD1, RNA (5'-R(*UP*AP*GP*UP*AP*AP*UP*C)-3') | | Authors: | Franco-Echevarria, E, Perez-Canadillas, J.M, Gonzalez, B. | | Deposit date: | 2017-05-19 | | Release date: | 2017-08-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.53 Å) | | Cite: | The structure of transcription termination factor Nrd1 reveals an original mode for GUAA recognition.
Nucleic Acids Res., 45, 2017
|
|
6HIT
 
 | | The crystal structure of haemoglobin from Atlantic cod | | Descriptor: | Hemoglobin alpha 2 chain, Hemoglobin beta 4 chain, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Helland, R, Bjorkeng, E.K, Rothweiler, U, Sydnes, M.O, Pampanin, D.M. | | Deposit date: | 2018-08-30 | | Release date: | 2019-08-21 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of haemoglobin from Atlantic cod.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
6NM3
 
 | | NMR structure of WW295 | | Descriptor: | WW295 peptide | | Authors: | Wang, G, Zarena, D. | | Deposit date: | 2019-01-10 | | Release date: | 2020-07-15 | | Last modified: | 2020-09-09 | | Method: | SOLUTION NMR | | Cite: | Two distinct amphipathic peptide antibiotics with systemic efficacy.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
5O1Y
 
 | | Structure of Nrd1 RNA binding domain in complex with RNA (GUAA) | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Protein NRD1, ... | | Authors: | Franco-Echevarria, E, Perez-Canadillas, J.M, Gonzalez, B. | | Deposit date: | 2017-05-19 | | Release date: | 2017-08-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | The structure of transcription termination factor Nrd1 reveals an original mode for GUAA recognition.
Nucleic Acids Res., 45, 2017
|
|
6CXQ
 
 | | HRFLRH peptide NMR structure in the presence of CO2 | | Descriptor: | Hexapeptide HRFLRH | | Authors: | Pires, D.A.T, Arake, L.M.R, Silva, L.P, Lopez-Castillo, A, Prates, M.V, Nascimento, C.J, Bloch Jr, C. | | Deposit date: | 2018-04-03 | | Release date: | 2018-07-04 | | Last modified: | 2024-10-09 | | Method: | SOLUTION NMR | | Cite: | A previously undescribed hexapeptide His-Arg-Phe-Leu-Arg-His-NH2from amphibian skin secretion shows CO2and metal biding affinities.
Peptides, 106, 2018
|
|
6CFK
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with D-histidyl-CAM and bound to protein Y (YfiA) at 2.7A resolution | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 16S Ribosomal RNA, 23S Ribosomal RNA, ... | | Authors: | Tereshchenkov, A.G, Dobosz-Bartoszek, M, Osterman, I.A, Marks, J, Sergeeva, V.A, Kasatsky, P, Komarova, E.S, Stavrianidi, A.N, Rodin, I.A, Konevega, A.L, Sergiev, P.V, Sumbatyan, N.V, Mankin, A.S, Bogdanov, A.A, Polikanov, Y.S. | | Deposit date: | 2018-02-15 | | Release date: | 2018-03-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Binding and Action of Amino Acid Analogs of Chloramphenicol upon the Bacterial Ribosome.
J. Mol. Biol., 430, 2018
|
|
5O1X
 
 | | Structure of Nrd1 RNA binding domain | | Descriptor: | 1,2-ETHANEDIOL, Protein NRD1, THIOCYANATE ION | | Authors: | Franco-Echevarria, E, Perez-Canadillas, J.M, Gonzalez, B. | | Deposit date: | 2017-05-19 | | Release date: | 2017-08-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The structure of transcription termination factor Nrd1 reveals an original mode for GUAA recognition.
Nucleic Acids Res., 45, 2017
|
|
6N60
 
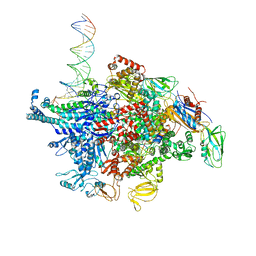 | | Escherichia coli RNA polymerase sigma70-holoenzyme bound to upstream fork promoter DNA and Microcin J25 (MccJ25) | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Braffman, N, Hauver, J, Campbell, E.A, Darst, S.A. | | Deposit date: | 2018-11-23 | | Release date: | 2019-01-09 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.68 Å) | | Cite: | Structural mechanism of transcription inhibition by lasso peptides microcin J25 and capistruin.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
6HK3
 
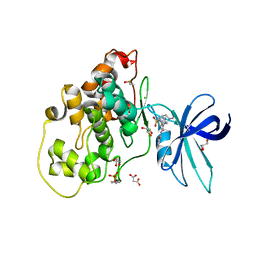 | | Crystal structure of GSK-3B in complex with pyrazine inhibitor C44 | | Descriptor: | 3-azanyl-~{N}-(2-methoxyphenyl)-6-[4-(4-methylpiperazin-1-yl)sulfonylphenyl]pyrazine-2-carboxamide, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Piretti, V, Giabbai, B, Demitri, N, Di Martino, R, Tripathi, S.K, Gobbo, D, Decherchi, S, Storici, P, Girotto, S, Cavalli, A. | | Deposit date: | 2018-09-05 | | Release date: | 2019-07-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Investigating Drug-Target Residence Time in Kinases through Enhanced Sampling Simulations.
J Chem Theory Comput, 15, 2019
|
|
