5TRF
 
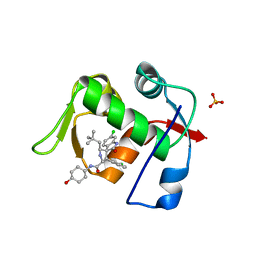 | | MDM2 in complex with SAR405838 | | Descriptor: | (2'S,3R,4'S,5'R)-6-chloro-4'-(3-chloro-2-fluorophenyl)-2'-(2,2-dimethylpropyl)-N-(trans-4-hydroxycyclohexyl)-2-oxo-1,2-dihydrospiro[indole-3,3'-pyrrolidine]-5'-carboxamide, E3 ubiquitin-protein ligase Mdm2, GLYCEROL, ... | | Authors: | Meagher, J.L, Stuckey, J.A. | | Deposit date: | 2016-10-26 | | Release date: | 2016-11-09 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | SAR405838: an optimized inhibitor of MDM2-p53 interaction that induces complete and durable tumor regression.
Cancer Res., 74, 2014
|
|
7L82
 
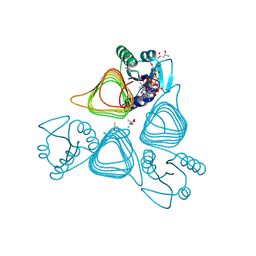 | | x-ray structure of the psychrobacter cryohalolentis Pcryo_0637 N-acetyltransferase in the presene of its reaction tetrahedral intermediate | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, Putative acetyl transferase protein, ... | | Authors: | Linehan, M.P, Thoden, J.B, Holden, H.M. | | Deposit date: | 2020-12-30 | | Release date: | 2021-03-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Characterization of two enzymes from Psychrobacter cryohalolentis that are required for the biosynthesis of an unusual diacetamido-d-sugar.
J.Biol.Chem., 296, 2021
|
|
4CJQ
 
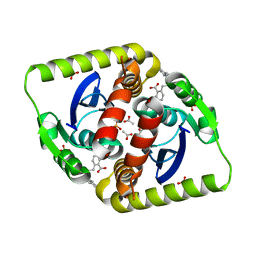 | | Interrogating HIV integrase for compounds that bind- a SAMPL challenge | | Descriptor: | 5-[(2S)-2-[[2-(1H-indol-3-yl)ethanoylamino]methyl]-4-methyl-pentyl]-1,3-benzodioxole-4-carboxylic acid, ACETATE ION, INTEGRASE, ... | | Authors: | Peat, T.S. | | Deposit date: | 2013-12-22 | | Release date: | 2014-01-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Interrogating HIV Integrase for Compounds that Bind- a Sampl Challenge.
J.Comput.Aided Mol.Des., 28, 2014
|
|
7FBT
 
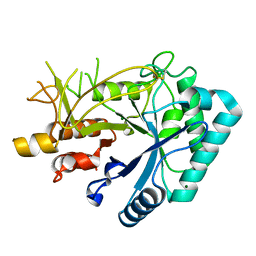 | | Crystal structure of chitinase (RmChi1) from Rhizomucor miehei (sp p32 2 1, MR) | | Descriptor: | Chitinase, MAGNESIUM ION | | Authors: | Jiang, Z.Q, Hu, S.Q, Zhu, Q, Liu, Y.C, Ma, J.W, Yan, Q.J, Gao, Y.G, Yang, S.Q. | | Deposit date: | 2021-07-12 | | Release date: | 2021-08-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a chitinase (RmChiA) from the thermophilic fungus Rhizomucor miehei with a real active site tunnel.
Biochim Biophys Acta Proteins Proteom, 1869, 2021
|
|
4CJ0
 
 | | Crystal structure of CelD in complex with affitin E12 | | Descriptor: | CALCIUM ION, E12 AFFITIN, ENDOGLUCANASE D, ... | | Authors: | Correa, A, Pacheco, S, Mechaly, A.E, Obal, G, Behar, G, Mouratou, B, Oppezzo, P, Alzari, P.M, Pecorari, F. | | Deposit date: | 2013-12-18 | | Release date: | 2014-05-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Potent and Specific Inhibition of Glycosidases by Small Artificial Binding Proteins (Affitins)
Plos One, 9, 2014
|
|
5X28
 
 | | Crystal structure of EGFR 696-1022 L858R in complex with SKLB(6) | | Descriptor: | 9-cyclohexyl-N2-[4-(4-methylpiperazin-1-yl)phenyl]-N8-phenyl-purine-2,8-diamine, CHLORIDE ION, Epidermal growth factor receptor | | Authors: | Yun, C.H. | | Deposit date: | 2017-01-31 | | Release date: | 2018-02-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.952 Å) | | Cite: | Structural insights into drug development strategy targeting EGFR T790M/C797S.
Oncotarget, 9, 2018
|
|
5U5P
 
 | | Crystal Structure and X-ray Diffraction Data Collection of Importin-alpha from Mus Musculus Complexed with a MLH1 NLS Peptide | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, DNA mismatch repair protein Mlh1, Importin subunit alpha-1 | | Authors: | Barros, A.C, Takeda, A.A, Dreyer, T.R, Velazquez-Campoy, A, Kobe, B, Fontes, M.R. | | Deposit date: | 2016-12-07 | | Release date: | 2018-03-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.171 Å) | | Cite: | DNA mismatch repair proteins MLH1 and PMS2 can be imported to the nucleus by a classical nuclear import pathway.
Biochimie, 146, 2018
|
|
3EAX
 
 | | Crystal structure PTP1B complex with small molecule compound LZP-6 | | Descriptor: | 4,4'-piperazine-1,4-diylbis{1-[3-(benzyloxy)phenyl]-4-oxobutane-1,3-dione}, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Zhang, Z.-Y, Liu, S, Zhang, L.-F, Yu, X, Xue, T, Gunawan, A.M, Long, Y.-Q. | | Deposit date: | 2008-08-26 | | Release date: | 2009-07-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Targeting inactive enzyme conformation: aryl diketoacid derivatives as a new class of PTP1B inhibitors.
J.Am.Chem.Soc., 130, 2008
|
|
5U7D
 
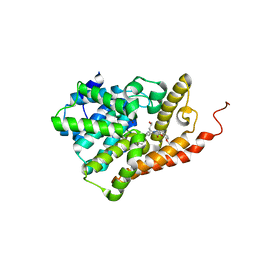 | | PDE2 catalytic domain complexed with inhibitors | | Descriptor: | 2-(3,4-dimethoxybenzyl)-7-[(2R,3R)-2-hydroxy-6-phenylhexan-3-yl]-5-methylimidazo[5,1-f][1,2,4]triazin-4(3H)-one, MAGNESIUM ION, ZINC ION, ... | | Authors: | Pandit, J, Parris, K. | | Deposit date: | 2016-12-12 | | Release date: | 2017-06-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Application of Structure-Based Design and Parallel Chemistry to Identify a Potent, Selective, and Brain Penetrant Phosphodiesterase 2A Inhibitor.
J. Med. Chem., 60, 2017
|
|
4CJ1
 
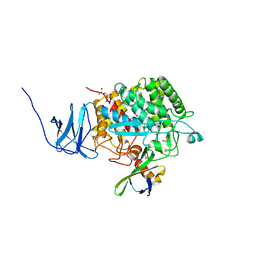 | | Crystal structure of CelD in complex with affitin H3 | | Descriptor: | CALCIUM ION, ENDOGLUCANASE D, GLYCEROL, ... | | Authors: | Correa, A, Pacheco, S, Mechaly, A.E, Obal, G, Behar, G, Mouratou, B, Oppezzo, P, Alzari, P.M, Pecorari, F. | | Deposit date: | 2013-12-18 | | Release date: | 2014-05-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Potent and Specific Inhibition of Glycosidases by Small Artificial Binding Proteins (Affitins)
Plos One, 9, 2014
|
|
4CJU
 
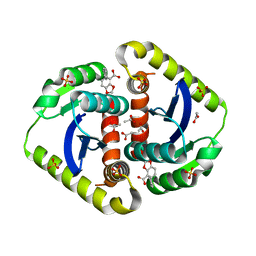 | |
4CKW
 
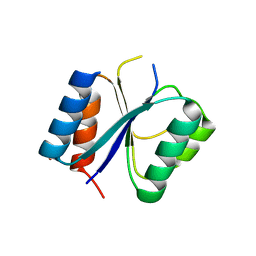 | | Structure of the Mycobacterium tuberculosis Type II Dehydroquinase N12S mutant (Crystal Form 1) | | Descriptor: | 3-DEHYDROQUINATE DEHYDRATASE, GLYCEROL | | Authors: | Otero, J.M, Llamas-Saiz, A.L, Maneiro, M, Peon, A, Sedes, A, Lamb, H, Hawkins, A.R, Gonzalez-Bello, C, van Raaij, M.J. | | Deposit date: | 2014-01-10 | | Release date: | 2015-03-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Investigation of the Dehydratation Mechanism Catalyzed by the Type II Dehydroquinase
To be Published
|
|
5UAB
 
 | |
3EB4
 
 | | Voltage-dependent K+ channel beta subunit (I211R) in complex with cortisone | | Descriptor: | 17,21-DIHYDROXYPREGNA-1,4-DIENE-3,11,20-TRIONE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Voltage-gated potassium channel subunit beta-2 | | Authors: | Pan, Y, Weng, J, Kabaleeswaran, V, Li, H, Cao, Y, Bhosle, R.C, Zhou, M. | | Deposit date: | 2008-08-26 | | Release date: | 2008-09-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Cortisone dissociates the Shaker family K+ channels from their beta subunits.
Nat.Chem.Biol., 4, 2008
|
|
2Y7C
 
 | | Atomic model of the Ocr-bound methylase complex from the Type I restriction-modification enzyme EcoKI (M2S1). Based on fitting into EM map 1534. | | Descriptor: | GENE 0.3 PROTEIN, TYPE I RESTRICTION ENZYME ECOKI M PROTEIN, TYPE-1 RESTRICTION ENZYME ECOKI SPECIFICITY PROTEIN | | Authors: | Kennaway, C.K, Obarska-Kosinska, A, White, J.H, Tuszynska, I, Cooper, L.P, Bujnicki, J.M, Trinick, J, Dryden, D.T.F. | | Deposit date: | 2011-01-31 | | Release date: | 2011-02-09 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (18 Å) | | Cite: | The Structure of M.Ecoki Type I DNA Methyltransferase with a DNA Mimic Antirestriction Protein.
Nucleic Acids Res., 37, 2009
|
|
5WBZ
 
 | | Structure of human Ketohexokinase complexed with hits from fragment screening | | Descriptor: | 6-[(3S,4S)-3,4-dihydroxypyrrolidin-1-yl]-2-[(3S)-3-hydroxy-3-methylpyrrolidin-1-yl]-4-(trifluoromethyl)pyridine-3-carbonitrile, CITRIC ACID, Ketohexokinase, ... | | Authors: | Pandit, J. | | Deposit date: | 2017-06-29 | | Release date: | 2017-09-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Discovery of Fragment-Derived Small Molecules for in Vivo Inhibition of Ketohexokinase (KHK).
J. Med. Chem., 60, 2017
|
|
5TQ6
 
 | |
5C2A
 
 | | PDE10 complexed with 6-chloro-2-cyclopropyl-N-[(2,4-dimethylthiazol-5-yl)methyl]-5-methyl-pyrimidin-4-amine | | Descriptor: | 6-chloro-2-cyclopropyl-N-[(2,4-dimethyl-1,3-thiazol-5-yl)methyl]-5-methylpyrimidin-4-amine, MAGNESIUM ION, ZINC ION, ... | | Authors: | Yan, Y. | | Deposit date: | 2015-06-15 | | Release date: | 2015-09-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery and Optimization of a Series of Pyrimidine-Based Phosphodiesterase 10A (PDE10A) Inhibitors through Fragment Screening, Structure-Based Design, and Parallel Synthesis.
J.Med.Chem., 58, 2015
|
|
3IXL
 
 | |
5UFE
 
 | | Wild-type K-Ras(GNP)/R11.1.6 complex | | Descriptor: | CADMIUM ION, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Parker, J.A, Mattos, C. | | Deposit date: | 2017-01-04 | | Release date: | 2017-08-02 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.302 Å) | | Cite: | An engineered protein antagonist of K-Ras/B-Raf interaction.
Sci Rep, 7, 2017
|
|
3S0O
 
 | | CDK2 in complex with inhibitor RC-1-138 | | Descriptor: | Cyclin-dependent kinase 2, [4-amino-2-(prop-2-en-1-ylamino)-1,3-thiazol-5-yl](pyridin-2-yl)methanone | | Authors: | Betzi, S, Alam, R, Han, H, Becker, A, Schonbrunn, E. | | Deposit date: | 2011-05-13 | | Release date: | 2012-10-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Development of highly potent and selective diaminothiazole inhibitors of cyclin-dependent kinases.
J.Med.Chem., 56, 2013
|
|
4I4F
 
 | | Structure of Focal Adhesion Kinase catalytic domain in complex with an allosteric binding pyrazolobenzothiazine compound. | | Descriptor: | Focal adhesion kinase 1, ISOPROPYL ALCOHOL, N-(4-tert-butylbenzyl)-1,5-dimethyl-1,5-dihydropyrazolo[4,3-c][2,1]benzothiazin-8-amine 4,4-dioxide | | Authors: | Skene, R.J, Hosfield, D.J. | | Deposit date: | 2012-11-27 | | Release date: | 2013-02-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure-based discovery of cellular-active allosteric inhibitors of FAK.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
6UDR
 
 | | S2 symmetric peptide design number 3 crystal form 1, Lurch | | Descriptor: | S2-3, Lurch crystal form 1 | | Authors: | Mulligan, V.K, Kang, C.S, Antselovich, I, Sawaya, M.R, Yeates, T.O, Baker, D. | | Deposit date: | 2019-09-19 | | Release date: | 2020-09-23 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Computational design of mixed chirality peptide macrocycles with internal symmetry.
Protein Sci., 29, 2020
|
|
4I6I
 
 | |
6MN1
 
 | | Crystal structure of meta-AAC0038, an environmental aminoglycoside resistance enzyme, mutant H168A in abortive complex with gentamicin-CoA | | Descriptor: | (2R,3R,4R,5R)-2-((1S,2S,3R,4S,6R)-4,6-DIAMINO-3-((2R,3R,6S)-3-AMINO-6-(AMINOMETHYL)-TETRAHYDRO-2H-PYRAN-2-YLOXY)-2-HYDR OXYCYCLOHEXYLOXY)-5-METHYL-4-(METHYLAMINO)-TETRAHYDRO-2H-PYRAN-3,5-DIOL, 3,6,9,12,15,18,21,24,27,30,33,36,39-TRIDECAOXAHENTETRACONTANE-1,41-DIOL, Aminoglycoside N(3)-acetyltransferase, ... | | Authors: | Stogios, P.J, Skarina, T, Michalska, K, Xu, Z, Yim, V, Savchenko, A, Joachimiak, A, Satchell, K.J, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-10-01 | | Release date: | 2018-10-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of meta-AAC0038, an environmental aminoglycoside resistance enzyme, mutant H168A in abortive complex with gentamicin-CoA
To Be Published
|
|
