5XOX
 
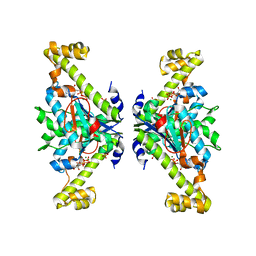 | | Crystal structure of tRNA(His) guanylyltranserase from Saccharomyces cerevisiae | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Lee, K, Lee, E.H, Son, J, Hwang, K.Y. | | Deposit date: | 2017-05-31 | | Release date: | 2017-07-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of tRNA(His) guanylyltransferase from Saccharomyces cerevisiae
Biochem. Biophys. Res. Commun., 490, 2017
|
|
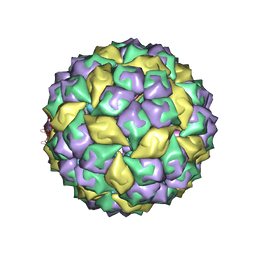
5ODV
 
 | | Structure of Watermelon mosaic virus potyvirus. | | Descriptor: | RNA (5'-R(P*UP*UP*UP*UP*U)-3'), coat protein | | Authors: | Zamora, M, Mendez-Lopez, E, Agirrezabala, X, Cuesta, R, Lavin, J.L, Sanchez-Pina, M.A, Aranda, M, Valle, M. | | Deposit date: | 2017-07-06 | | Release date: | 2017-09-27 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Potyvirus virion structure shows conserved protein fold and RNA binding site in ssRNA viruses.
Sci Adv, 3, 2017
|
|
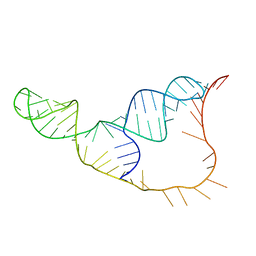
2B2E
 
 | | RNA stemloop from bacteriophage MS2 complexed with an N87S,E89K mutant MS2 capsid | | Descriptor: | 5'-R(*AP*CP*AP*UP*GP*AP*GP*GP*AP*UP*UP*AP*CP*CP*CP*AP*UP*GP*U)-3', Coat protein | | Authors: | Horn, W.T, Tars, K, Grahn, E, Helgstrand, C, Baron, A.J, Lago, H, Adams, C.J, Peabody, D.S, Phillips, S.E.V, Stonehouse, N.J, Liljas, L, Stockley, P.G. | | Deposit date: | 2005-09-19 | | Release date: | 2006-05-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Structural basis of RNA binding discrimination between bacteriophages Qbeta and MS2
Structure, 14, 2006
|
|
2N8V
 
 | | An NMR/SAXS structure of the PKI domain of the honeybee dicistrovirus, Israeli acute paralysis virus (IAPV) IRES | | Descriptor: | RNA (70-MER) | | Authors: | Au, H.H, Cornilescu, G, Mouzakis, K.D, Burke, J.E, Ren, Q, Lee, S, Butcher, S.E, Jan, E. | | Deposit date: | 2015-10-27 | | Release date: | 2015-11-11 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Global shape mimicry of tRNA within a viral internal ribosome entry site mediates translational reading frame selection.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
2RS2
 
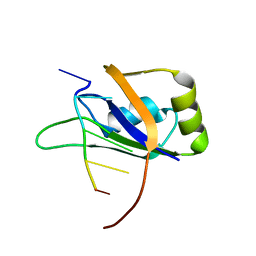 | | 1H, 13C, and 15N Chemical Shift Assignments for Musashi1 RBD1:r(GUAGU) complex | | Descriptor: | RNA (5'-R(*GP*UP*AP*GP*U)-3'), RNA-binding protein Musashi homolog 1 | | Authors: | Ohyama, T, Nagata, T, Tsuda, K, Imai, T, Okano, H, Yamazaki, T, Katahira, M. | | Deposit date: | 2011-06-27 | | Release date: | 2011-12-28 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of Musashi1 in a complex with target RNA: the role of aromatic stacking interactions
Nucleic Acids Res., 2011
|
|
7EB1
 
 | | Solution NMR structure of the RRM domain of RNA binding protein RBM3 from homo sapiens | | Descriptor: | RNA-binding protein 3 | | Authors: | Boral, S, Roy, S, Basak, A.J, Maiti, S, Lee, W, De, S. | | Deposit date: | 2021-03-08 | | Release date: | 2021-12-08 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural and dynamic studies of the human RNA binding protein RBM3 reveals the molecular basis of its oligomerization and RNA recognition.
Febs J., 289, 2022
|
|
2KF6
 
 | |
2KF4
 
 | | Barnase high pressure structure | | Descriptor: | Ribonuclease | | Authors: | Williamson, M.P, Wilton, D.J. | | Deposit date: | 2009-02-11 | | Release date: | 2009-12-08 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Pressure-dependent structure changes in barnase on ligand binding reveal intermediate rate fluctuations.
Biophys.J., 97, 2009
|
|
2KF3
 
 | |
2KF5
 
 | |
1HBX
 
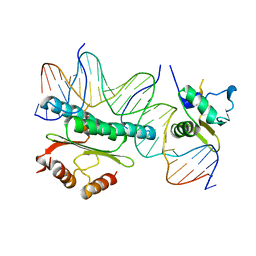 | | Ternary Complex of SAP-1 and SRF with specific SRE DNA | | Descriptor: | 5'-D(*CP*AP*CP*AP*CP*CP*GP*GP*AP*AP*GP*TP*CP* CP*TP*AP*AP*TP*TP*AP*GP*GP*CP*CP*AP*T)-3', 5'-D(*GP*AP*TP*GP*GP*CP*CP*TP*AP*AP*TP*TP*AP* GP*GP*AP*CP*TP*TP*CP*CP*GP*GP*TP*G)-3', ETS-DOMAIN PROTEIN ELK-4, ... | | Authors: | Hassler, M, Richmond, T.J. | | Deposit date: | 2001-04-20 | | Release date: | 2001-06-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | The B-Box Dominates Sap-1/Srf Interactions in the Structure of the Ternary Complex
Embo J., 20, 2001
|
|
6YFT
 
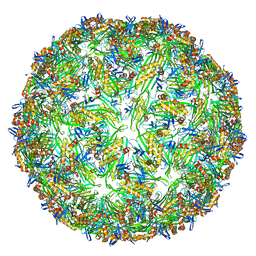 | | Virus-like particle of Wenzhou levi-like virus 1 | | Descriptor: | RNA, coat protein | | Authors: | Rumnieks, J, Kalnins, G, Sisovs, M, Lieknina, I, Tars, K. | | Deposit date: | 2020-03-26 | | Release date: | 2020-09-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Three-dimensional structure of 22 uncultured ssRNA bacteriophages: Flexibility of the coat protein fold and variations in particle shapes.
Sci Adv, 6, 2020
|
|
4B8T
 
 | | RNA BINDING PROTEIN Solution structure of the third KH domain of KSRP in complex with the G-rich target sequence. | | Descriptor: | 5'-R(*AP*GP*GP*GP*UP)-3', KH-TYPE SPLICING REGULATORY PROTEIN | | Authors: | Nicastro, G, Garcia-Mayoral, M.F, Hollingworth, D, Kelly, G, Martin, S.R, Briata, P, Gherzi, R, Ramos, A. | | Deposit date: | 2012-08-30 | | Release date: | 2012-11-07 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Noncanonical G Recognition Mediates Ksrp Regulation of Let-7 Biogenesis
Nat.Struct.Mol.Biol., 19, 2012
|
|
1SGV
 
 | | STRUCTURE OF TRNA PSI55 PSEUDOURIDINE SYNTHASE (TRUB) | | Descriptor: | tRNA pseudouridine synthase B | | Authors: | Chaudhuri, B.N, Chan, S, Perry, L.J, Yeates, T.O, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2004-02-24 | | Release date: | 2004-03-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the apo forms of psi 55 tRNA pseudouridine synthase from Mycobacterium tuberculosis: a hinge at the base of the catalytic cleft.
J.Biol.Chem., 279, 2004
|
|
4JIY
 
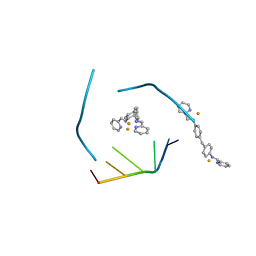 | | RNA three-way junction stabilized by a supramolecular di-iron(II) cylinder drug | | Descriptor: | 5'-(CGUACG)-3', FE (II) ION, N-[(1E)-PYRIDIN-2-YLMETHYLENE]-N-[4-(4-{[(1E)-PYRIDIN-2-YLMETHYLENE]AMINO}BENZYL)PHENYL]AMINE | | Authors: | Sigel, R.K.O, Schnabl, J.A, Freisinger, E, Spingler, B, Hannon, M.J. | | Deposit date: | 2013-03-07 | | Release date: | 2013-09-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Binding of a designed anti-cancer drug to the central cavity of an RNA three-way junction.
Angew.Chem.Int.Ed.Engl., 52, 2013
|
|
4C7Q
 
 | |
7C7L
 
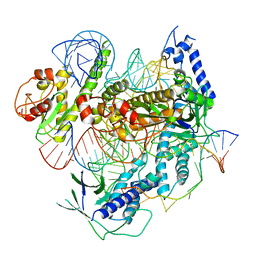 | | Cryo-EM structure of the Cas12f1-sgRNA-target DNA complex | | Descriptor: | CRISPR-associated protein Cas14a.1, DNA (40-mer), ZINC ION, ... | | Authors: | Takeda, N.S, Nakagawa, R, Okazaki, S, Hirano, H, Kobayashi, K, Kusakizako, T, Nishizawa, T, Yamashita, K, Nishimasu, H, Nureki, O. | | Deposit date: | 2020-05-26 | | Release date: | 2020-12-23 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of the miniature type V-F CRISPR-Cas effector enzyme.
Mol.Cell, 81, 2021
|
|
7DA7
 
 | |
7DAS
 
 | |
3NR5
 
 | | Crystal structure of human Maf1 | | Descriptor: | Repressor of RNA polymerase III transcription MAF1 homolog | | Authors: | Ringel, R, Vannini, A, Kusser, A.G, Berninghausen, O, Kassavetis, G.A, Cramer, P. | | Deposit date: | 2010-06-30 | | Release date: | 2010-10-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Molecular Basis of RNA Polymerase III Transcription Repression by Maf1
Cell(Cambridge,Mass.), 143, 2010
|
|
2HCS
 
 | |
4MTP
 
 | | RdRp from Japanesese Encephalitis Virus | | Descriptor: | RNA dependent RNA polymerase, ZINC ION | | Authors: | Surana, P, Nair, D.T. | | Deposit date: | 2013-09-20 | | Release date: | 2013-12-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.65 Å) | | Cite: | RNA-dependent RNA polymerase of Japanese encephalitis virus binds the initiator nucleotide GTP to form a mechanistically important pre-initiation state.
Nucleic Acids Res., 42, 2014
|
|
2H49
 
 | |
1HHT
 
 | | RNA dependent RNA polymerase from dsRNA bacteriophage phi6 plus template | | Descriptor: | DNA (5'-(*TP*TP*TP*CP*C)-3'), MANGANESE (II) ION, P2 PROTEIN | | Authors: | Grimes, J.M, Butcher, S.J, Makeyev, E.V, Bamford, D.H, Stuart, D.I. | | Deposit date: | 2000-12-28 | | Release date: | 2001-03-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A Mechanism for Initiating RNA-Dependent RNA Polymerization
Nature, 410, 2001
|
|
7Q48
 
 | |
