5QDJ
 
 | | PanDDA analysis group deposition -- Crystal structure of PTP1B in complex with compound_FMOPL000211a | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, N-(4-methyl-1,3-thiazol-2-yl)propanamide, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Keedy, D.A, Hill, Z.B, Biel, J.T, Kang, E, Rettenmaier, T.J, Brandao-Neto, J, von Delft, F, Wells, J.A, Fraser, J.S. | | Deposit date: | 2018-08-30 | | Release date: | 2018-10-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.761 Å) | | Cite: | An expanded allosteric network in PTP1B by multitemperature crystallography, fragment screening, and covalent tethering.
Elife, 7, 2018
|
|
3PW4
 
 | |
3FU3
 
 | |
5QDX
 
 | | PanDDA analysis group deposition -- Crystal structure of PTP1B in complex with compound_FMOPL000484a | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Tyrosine-protein phosphatase non-receptor type 1, {N}-[(2~{S})-1-diazanyl-3-(4-hydroxyphenyl)-1-oxidanylidene-propan-2-yl]ethanamide | | Authors: | Keedy, D.A, Hill, Z.B, Biel, J.T, Kang, E, Rettenmaier, T.J, Brandao-Neto, J, von Delft, F, Wells, J.A, Fraser, J.S. | | Deposit date: | 2018-08-30 | | Release date: | 2018-10-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.062 Å) | | Cite: | An expanded allosteric network in PTP1B by multitemperature crystallography, fragment screening, and covalent tethering.
Elife, 7, 2018
|
|
5QEE
 
 | | PanDDA analysis group deposition -- Crystal structure of PTP1B in complex with compound_FMOPL000240a | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-cyclopentyl-N-(5-methyl-1,3-thiazol-2-yl)propanamide, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Keedy, D.A, Hill, Z.B, Biel, J.T, Kang, E, Rettenmaier, T.J, Brandao-Neto, J, von Delft, F, Wells, J.A, Fraser, J.S. | | Deposit date: | 2018-08-30 | | Release date: | 2018-10-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.932 Å) | | Cite: | An expanded allosteric network in PTP1B by multitemperature crystallography, fragment screening, and covalent tethering.
Elife, 7, 2018
|
|
5QEP
 
 | | PanDDA analysis group deposition -- Crystal structure of PTP1B in complex with compound_XST00000692b | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-[4-(trifluoromethyl)phenyl]-1,3-thiazole-4-carboxylic acid, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Keedy, D.A, Hill, Z.B, Biel, J.T, Kang, E, Rettenmaier, T.J, Brandao-Neto, J, von Delft, F, Wells, J.A, Fraser, J.S. | | Deposit date: | 2018-08-30 | | Release date: | 2018-10-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.774 Å) | | Cite: | An expanded allosteric network in PTP1B by multitemperature crystallography, fragment screening, and covalent tethering.
Elife, 7, 2018
|
|
4ACH
 
 | | GSK3b in complex with inhibitor | | Descriptor: | 3-AMINO-N-(3-METHOXYPROPYL)-6-{4-[(4-METHYLPIPERAZIN-1-YL)SULFONYL]PHENYL}PYRAZINE-2-CARBOXAMIDE, GLYCOGEN SYNTHASE KINASE-3 BETA | | Authors: | Xue, Y, Ormo, M. | | Deposit date: | 2011-12-15 | | Release date: | 2012-05-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Discovery of novel potent and highly selective glycogen synthase kinase-3 beta (GSK3 beta ) inhibitors for Alzheimer's disease: design, synthesis, and characterization of pyrazines.
J. Med. Chem., 55, 2012
|
|
5QFI
 
 | | PanDDA analysis group deposition -- Crystal structure of PTP1B in complex with compound_FMOOA000531a | | Descriptor: | (1R,4R,5R,6S)-4,6-dihydroxy-N-phenyl-2-azabicyclo[3.3.1]nonane-2-carboxamide, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Keedy, D.A, Hill, Z.B, Biel, J.T, Kang, E, Rettenmaier, T.J, Brandao-Neto, J, von Delft, F, Wells, J.A, Fraser, J.S. | | Deposit date: | 2018-08-30 | | Release date: | 2018-10-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | An expanded allosteric network in PTP1B by multitemperature crystallography, fragment screening, and covalent tethering.
Elife, 7, 2018
|
|
4ACD
 
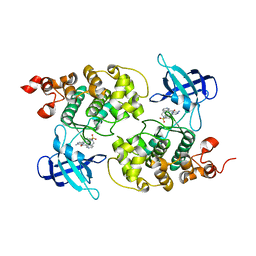 | | GSK3b in complex with inhibitor | | Descriptor: | 3-AMINO-6-{4-[(4-METHYLPIPERAZIN-1-YL)SULFONYL]PHENYL}-N-PYRIDIN-3-YLPYRAZINE-2-CARBOXAMIDE, GLYCOGEN SYNTHASE KINASE-3 BETA | | Authors: | Xue, Y, Ormo, M. | | Deposit date: | 2011-12-15 | | Release date: | 2012-05-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Discovery of novel potent and highly selective glycogen synthase kinase-3 beta (GSK3 beta ) inhibitors for Alzheimer's disease: design, synthesis, and characterization of pyrazines.
J. Med. Chem., 55, 2012
|
|
4RIA
 
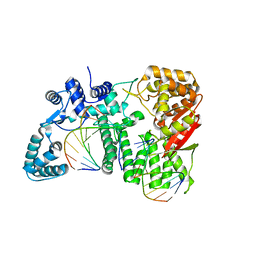 | | FAN1 Nuclease bound to 5' phosphorylated nicked DNA | | Descriptor: | BARIUM ION, DNA (5'-D(*TP*TP*TP*TP*TP*TP*G*AP*GP*GP*CP*GP*TP*G)-3'), DNA (5'-D(P*AP*GP*AP*CP*TP*CP*CP*TP*C)-3'), ... | | Authors: | Pavletich, N.P, Wang, R. | | Deposit date: | 2014-10-05 | | Release date: | 2014-12-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | DNA repair. Mechanism of DNA interstrand cross-link processing by repair nuclease FAN1.
Science, 346, 2014
|
|
4RID
 
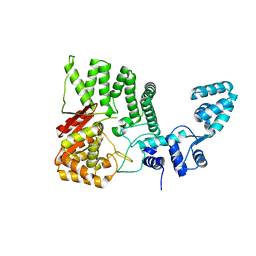 | | Human FAN1 nuclease | | Descriptor: | Fanconi-associated nuclease 1 | | Authors: | Pavletich, N.P, Wang, R. | | Deposit date: | 2014-10-05 | | Release date: | 2014-12-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | DNA repair. Mechanism of DNA interstrand cross-link processing by repair nuclease FAN1.
Science, 346, 2014
|
|
5WCX
 
 | |
4A7K
 
 | | Bifunctional Aldos-2-ulose dehydratase | | Descriptor: | ALDOS-2-ULOSE DEHYDRATASE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Claesson, M, Lindqvist, Y, Madrid, S, Sandalova, T, Fiskesund, R, Yu, S, Schneider, G. | | Deposit date: | 2011-11-14 | | Release date: | 2012-03-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Bifunctional Aldos-2-Ulose Dehydratase/Isomerase from Phanerochaete Chrysosporium with the Reaction Intermediate Ascopyrone M.
J.Mol.Biol., 417, 2012
|
|
5DC7
 
 | | Crystal structure of D176A-Y306F HDAC8 in complex with a tetrapeptide substrate | | Descriptor: | Fluor-de-Lys tetrapeptide assay substrate, GLYCEROL, Histone deacetylase 8, ... | | Authors: | Decroos, C, Lee, M.S, Christianson, D.W. | | Deposit date: | 2015-08-23 | | Release date: | 2016-02-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | General Base-General Acid Catalysis in Human Histone Deacetylase 8.
Biochemistry, 55, 2016
|
|
2AJ3
 
 | | Crystal Structure of a Cross-Reactive HIV-1 Neutralizing CD4-Binding Site Antibody Fab m18 | | Descriptor: | Fab m18, Heavy Chain, Light Chain, ... | | Authors: | Prabakaran, P, Gan, J, Wu, Y.Q, Zhang, M.Y, Dimitrov, D.S, Ji, X. | | Deposit date: | 2005-08-01 | | Release date: | 2006-03-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structural mimicry of CD4 by a cross-reactive HIV-1 neutralizing antibody with CDR-H2 and H3 containing unique motifs.
J.Mol.Biol., 357, 2006
|
|
4REA
 
 | | A Nuclease DNA complex | | Descriptor: | DNA (5'-D(*TP*GP*CP*TP*CP*GP*CP*CP*AP*C)-3'), DNA (5'-D(P*CP*GP*TP*GP*GP*CP*GP*AP*GP*C)-3'), DNA (5'-D(P*GP*GP*CP*GP*AP*GP*CP*GP*CP*TP*CP*GP*CP*CP*AP*CP*G)-3'), ... | | Authors: | Zhao, Q, Xue, X, Longerich, S, Sung, P, Xiong, Y. | | Deposit date: | 2014-09-22 | | Release date: | 2014-12-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.81 Å) | | Cite: | Structural insights into 5' flap DNA unwinding and incision by the human FAN1 dimer.
Nat Commun, 5, 2014
|
|
2X1M
 
 | | Crystal structure of Mycobacterium smegmatis methionyl-tRNA synthetase in complex with methionine | | Descriptor: | 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, DIHYDROGENPHOSPHATE ION, METHIONINE, ... | | Authors: | Ingvarsson, H, Jones, T.A, Unge, T. | | Deposit date: | 2009-12-31 | | Release date: | 2010-07-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Flexibility and Communication within the Structure of the Mycobacterium Smegmatis Methionyl-tRNA Synthetase.
FEBS J., 277, 2010
|
|
2X87
 
 | | Crystal Structure of the reconstituted CotA | | Descriptor: | 1,2-ETHANEDIOL, COPPER (II) ION, HYDROXIDE ION, ... | | Authors: | Bento, I, Silva, C.S, Chen, Z, Martins, L.O, Lindley, P.F, Soares, C.M. | | Deposit date: | 2010-03-06 | | Release date: | 2010-09-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mechanisms Underlying Dioxygen Reduction in Laccases. Structural and Modelling Studies Focusing on Proton Transfer.
Bmc Struct.Biol., 10, 2010
|
|
5D8W
 
 | | Structrue of a lucidum protein | | Descriptor: | Endoglucanase | | Authors: | Guo, R, Li, Q, Shang, N, Liu, G, Ko, T.P, Chen, C.C, Liu, W. | | Deposit date: | 2015-08-18 | | Release date: | 2016-06-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Functional and structural analyses of a 1,4-beta-endoglucanase from Ganoderma lucidum.
Enzyme.Microb.Technol., 86, 2016
|
|
7FP0
 
 | | PanDDA analysis group deposition -- Aar2/RNaseH in complex with fragment P08F02 from the F2X-Universal Library | | Descriptor: | 1-cyclopropylimidazolidin-2-one, A1 cistron-splicing factor AAR2, Pre-mRNA-splicing factor 8 | | Authors: | Barthel, T, Wollenhaupt, J, Lima, G.M.A, Wahl, M.C, Weiss, M.S. | | Deposit date: | 2022-08-26 | | Release date: | 2022-11-02 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Large-Scale Crystallographic Fragment Screening Expedites Compound Optimization and Identifies Putative Protein-Protein Interaction Sites.
J.Med.Chem., 65, 2022
|
|
5D45
 
 | | Crystal Structure of FABP4 in complex with 3-(5-cyclopropyl-2,3-diphenyl-1H-indol-1-yl)propanoic acid | | Descriptor: | 3-(5-cyclopropyl-2,3-diphenyl-1H-indol-1-yl)propanoic acid, Fatty acid-binding protein, adipocyte | | Authors: | Tagami, U, Takahashi, K, Igarashi, S, Ejima, C, Yoshida, T, Takeshita, S, Miyanaga, W, Sugiki, M, Tokumasu, M, Hatanaka, T, Kashiwagi, T, Ishikawa, K, Miyano, H, Mizukoshi, T. | | Deposit date: | 2015-08-07 | | Release date: | 2016-06-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Interaction Analysis of FABP4 Inhibitors by X-ray Crystallography and Fragment Molecular Orbital Analysis
Acs Med.Chem.Lett., 7, 2016
|
|
2X40
 
 | |
5TH3
 
 | |
4RGX
 
 | | Crystal Structure of Putative MarR Family Transcriptional Regulator HcaR from Acinetobacter sp. ADP complexed with 3,4-dihydroxy bezoic acid | | Descriptor: | 1,2-ETHANEDIOL, 3,4-DIHYDROXYBENZOIC ACID, CHLORIDE ION, ... | | Authors: | Kim, Y, Joachimiak, G, Bigelow, L, Cobb, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-09-30 | | Release date: | 2015-03-04 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.102 Å) | | Cite: | Crystal Structure of Putative MarR Family Transcriptional Regulator HcaR from Acinetobacter sp. ADP complexed with 3,4-dihydroxy bezoic acid
To be Published, 2014
|
|
3QXP
 
 | | CDK2 in complex with inhibitor RC-3-89 | | Descriptor: | 1,2-ETHANEDIOL, 4-{[4-amino-5-(2-nitrobenzoyl)-1,3-thiazol-2-yl]amino}benzenesulfonamide, Cyclin-dependent kinase 2 | | Authors: | Betzi, S, Alam, R, Han, H, Becker, A, Schonbrunn, E. | | Deposit date: | 2011-03-02 | | Release date: | 2012-10-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Development of highly potent and selective diaminothiazole inhibitors of cyclin-dependent kinases.
J.Med.Chem., 56, 2013
|
|
