5KAG
 
 | | Crystal structure of a dioxygenase in the Crotonase superfamily in P21 | | Descriptor: | (3,5-dihydroxyphenyl)acetyl-CoA 1,2-dioxygenase, OXYGEN MOLECULE, [(2R,3S,4R,5R)-5-(6-AMINO-9H-PURIN-9-YL)-4-HYDROXY-3-(PHOSPHONOOXY)TETRAHYDROFURAN-2-YL]METHYL (3R)-4-({3-[(2-{[(3,5-DIHYDROXYPHENYL)ACETYL]AMINO}ETHYL)AMINO]-3-OXOPROPYL}AMINO)-3-HYDROXY-2,2-DIMETHYL-4-OXOBUTYL DIHYDROGEN DIPHOSPHATE | | Authors: | Li, K, Fielding, E.N, Condurso, H.L, Bruner, S.D. | | Deposit date: | 2016-06-01 | | Release date: | 2017-06-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.456 Å) | | Cite: | Probing the structural basis of oxygen binding in a cofactor-independent dioxygenase.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
3HH1
 
 | | The Structure of a Tetrapyrrole methylase family protein domain from Chlorobium tepidum TLS | | Descriptor: | 1,2-ETHANEDIOL, SULFATE ION, Tetrapyrrole methylase family protein | | Authors: | Cuff, M.E, Sather, A, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-05-14 | | Release date: | 2009-07-07 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The Structure of a Tetrapyrrole methylase family protein domain from Chlorobium tepidum TLS.
TO BE PUBLISHED
|
|
5VN9
 
 | | Structure of bacteriorhodopsin from crystals grown at 4 deg C using GlyNCOC15+4 as an LCP host lipid | | Descriptor: | Bacteriorhodopsin | | Authors: | Ishchenko, A, Peng, L, Zinovev, E, Vlasov, A, Lee, S.C, Kuklin, A, Mishin, A, Borshchevskiy, V, Zhang, Q, Cherezov, V. | | Deposit date: | 2017-04-28 | | Release date: | 2017-07-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.594 Å) | | Cite: | Chemically Stable Lipids for Membrane Protein Crystallization.
Cryst Growth Des, 17, 2017
|
|
3HGJ
 
 | | Old Yellow Enzyme from Thermus scotoductus SA-01 complexed with p-hydroxy-benzaldehyde | | Descriptor: | Chromate reductase, FLAVIN MONONUCLEOTIDE, P-HYDROXYBENZALDEHYDE | | Authors: | Opperman, D.J, Sewell, B.T, Litthauer, D, Isupov, M.N, Littlechild, J.A, van Heerden, E. | | Deposit date: | 2009-05-14 | | Release date: | 2010-02-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a thermostable old yellow enzyme from Thermus scotoductus SA-01
Biochem.Biophys.Res.Commun., 393, 2010
|
|
2Y3P
 
 | | Crystal structure of N-terminal domain of GyrA with the antibiotic simocyclinone D8 | | Descriptor: | DNA GYRASE SUBUNIT A, MAGNESIUM ION, SIMOCYCLINONE D8 | | Authors: | Edwards, M.J, Flatman, R.H, Mitchenall, L.A, Stevenson, C.E.M, Le, T.B.K, Clarke, T.A, McKay, A.R, Fiedler, H.-P, Buttner, M.J, Lawson, D.M, Maxwell, A. | | Deposit date: | 2010-12-22 | | Release date: | 2010-12-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | A Crystal Structure of the Bifunctional Antibiotic Simocyclinone D8, Bound to DNA Gyrase.
Science, 326, 2009
|
|
2Y4G
 
 | | Structure of the Tirandamycin-bound FAD-dependent tirandamycin oxidase TamL in P212121 space group | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Carlson, J.C, Li, S, Gunatilleke, S.S, Anzai, Y, Burr, D.A, Podust, L.M, Sherman, D.H. | | Deposit date: | 2011-01-05 | | Release date: | 2011-06-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Tirandamycin Biosynthesis is Mediated by Co-Dependent Oxidative Enzymes
Nat.Chem, 3, 2011
|
|
3HDA
 
 | | PrtC methionine mutants: M226A_DESY | | Descriptor: | CALCIUM ION, CHLORIDE ION, Secreted protease C, ... | | Authors: | Oberholzer, A.E, Bumann, M, Hege, T, Russo, S, Baumann, U. | | Deposit date: | 2009-05-07 | | Release date: | 2009-06-30 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.131 Å) | | Cite: | Metzincin's canonical methionine is responsible for the structural integrity of the zinc-binding site
Biol.Chem., 390, 2009
|
|
2Y1Z
 
 | | Human alphaB Crystallin ACD R120G | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, ALPHA-CRYSTALLIN B CHAIN | | Authors: | Clark, A.R, Bagneris, C, Naylor, C.E, Keep, N.H, Slingsby, C. | | Deposit date: | 2010-12-10 | | Release date: | 2011-03-02 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of R120G Disease Mutant of Human Alphab-Crystallin Domain Dimer Shows Closure of a Groove
J.Mol.Biol., 408, 2011
|
|
5KDS
 
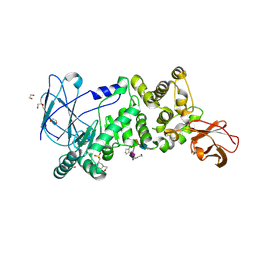 | | ZmpB metallopeptidase in complex with an O-glycopeptide (a2,6-sialylated core-3 pentapeptide). | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-[N-acetyl-alpha-neuraminic acid-(2-6)]2-acetamido-2-deoxy-alpha-D-galactopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Noach, I, Ficko-Blean, E, Stuart, C, Boraston, A.B. | | Deposit date: | 2016-06-08 | | Release date: | 2017-01-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Recognition of protein-linked glycans as a determinant of peptidase activity.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
2XGY
 
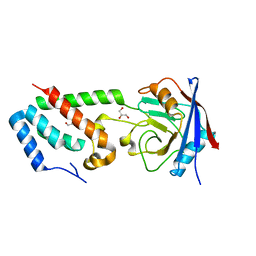 | | Complex of Rabbit Endogenous Lentivirus (RELIK)Capsid with Cyclophilin A | | Descriptor: | GLYCEROL, PEPTIDYL-PROLYL CIS-TRANS ISOMERASE A, RELIK CAPSID N-TERMINAL DOMAIN | | Authors: | Goldstone, D.C, Robertson, L.E, Haire, L.F, Stoye, J.P, Taylor, I.A. | | Deposit date: | 2010-06-08 | | Release date: | 2010-09-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and Functional Analysis of Prehistoric Lentiviruses Uncovers an Ancient Molecular Interface.
Cell Host Microbe, 8, 2010
|
|
3HDD
 
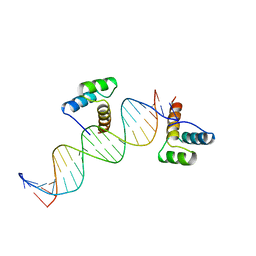 | | ENGRAILED HOMEODOMAIN DNA COMPLEX | | Descriptor: | 5'-D(*AP*TP*TP*AP*GP*GP*TP*AP*AP*TP*TP*AP*CP*AP*TP*GP*GP*CP*AP*AP*A)-3', 5'-D(*TP*TP*TP*TP*GP*CP*CP*AP*TP*GP*TP*AP*AP*TP*TP*AP*CP*CP*TP*AP*A)-3', ENGRAILED HOMEODOMAIN | | Authors: | Fraenkel, E, Rould, M.A, Chambers, K.A, Pabo, C.O. | | Deposit date: | 1998-07-13 | | Release date: | 1998-11-11 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Engrailed homeodomain-DNA complex at 2.2 A resolution: a detailed view of the interface and comparison with other engrailed structures.
J.Mol.Biol., 284, 1998
|
|
3HF3
 
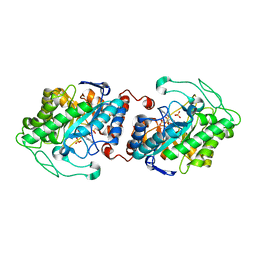 | | Old Yellow Enzyme from Thermus scotoductus SA-01 | | Descriptor: | Chromate reductase, FLAVIN MONONUCLEOTIDE, SULFATE ION | | Authors: | Opperman, D.J, Sewell, B.T, Litthauer, D, Isupov, M.N, Littlechild, J.A, van Heerden, E. | | Deposit date: | 2009-05-11 | | Release date: | 2010-02-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a thermostable old yellow enzyme from Thermus scotoductus SA-01
Biochem.Biophys.Res.Commun., 393, 2010
|
|
3HEJ
 
 | | Crystal structure of Staphylococcal nuclease variant Delta+PHS T62R at cryogenic temperature | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, GLYCEROL, ... | | Authors: | Khangulov, V.S, Schlessman, J.L, Heroux, A, Garcia-Moreno, E.B. | | Deposit date: | 2009-05-08 | | Release date: | 2010-05-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Domain swapping promoted by a single mutation that introduces an ionizable group into the hydrophobic core of a protein
To be Published
|
|
2YPG
 
 | | Haemagglutinin of 1968 Human H3N2 Virus in Complex with Human Receptor Analogue LSTc | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Liu, J, Xiong, X, Haire, L.F, Lin, Y.P, Wharton, S.A, Martin, S.R, Coombs, P.J, Vachieri, S.G, Christodoulou, E, Walker, P.A, Skehel, J.J, Gamblin, S.J, Hay, A.J, Daniels, R.S, McCauley, J.W. | | Deposit date: | 2012-10-30 | | Release date: | 2012-11-07 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Evolution of the Receptor Binding Properties of the Influenza A(H3N2) Hemagglutinin.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3HFQ
 
 | | Crystal structure of the lp_2219 protein from Lactobacillus plantarum. Northeast Structural Genomics Consortium Target LpR118. | | Descriptor: | PHOSPHATE ION, uncharacterized protein lp_2219 | | Authors: | Vorobiev, S.M, Scott, L, Schauder, C, Xiao, R, Ciccosanti, C, Foote, E.L, Maglaqui, M, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-05-12 | | Release date: | 2009-05-26 | | Last modified: | 2021-10-13 | | Method: | X-RAY DIFFRACTION (1.963 Å) | | Cite: | Crystal structure of the lp_2219 protein from Lactobacillus plantarum.
To be Published
|
|
3HGM
 
 | |
3HJ3
 
 | | Crystal Structure of the ChTS-DHFR F207A Non-Active Site Mutant | | Descriptor: | 10-PROPARGYL-5,8-DIDEAZAFOLIC ACID, 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, Chain A, ... | | Authors: | Anderson, K.S, Martucci, W.E. | | Deposit date: | 2009-05-20 | | Release date: | 2010-06-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Exploring novel strategies for AIDS protozoal pathogens: alpha-helix mimetics targeting a key allosteric protein-protein interaction in C. hominis TS-DHFR.
Medchemcomm, 4, 2013
|
|
5KRL
 
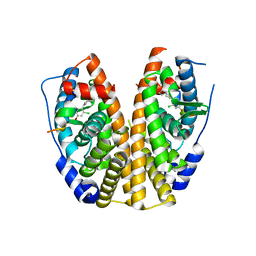 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in Complex with the A-CD ring estrogen, (1S,7aS)-5-(2-chloro-4-hydroxyphenyl)-7a-methyl-2,3,3a,4,7,7a-hexahydro-1H-inden-1-ol | | Descriptor: | (1~{S},3~{a}~{R},7~{a}~{S})-5-(2-chloranyl-4-oxidanyl-phenyl)-2,3,3~{a},4,7,7~{a}-hexahydro-1~{H}-inden-1-ol, Estrogen receptor, NCOA2 | | Authors: | Nwachukwu, J.C, Srinivasan, S, Bruno, N.E, Nowak, J, Kojetin, D.J, Elemento, O, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2016-07-07 | | Release date: | 2017-01-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Systems Structural Biology Analysis of Ligand Effects on ER alpha Predicts Cellular Response to Environmental Estrogens and Anti-hormone Therapies.
Cell Chem Biol, 24, 2017
|
|
5KUT
 
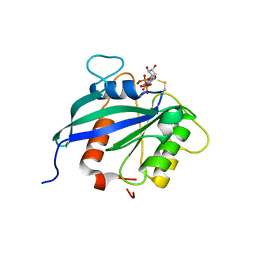 | | hMiro2 C-terminal GTPase domain, GDP-bound | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Mitochondrial Rho GTPase 2 | | Authors: | Klosowiak, J.L, Focia, P.J, Rice, S.E, Freymann, D.M. | | Deposit date: | 2016-07-13 | | Release date: | 2016-09-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.693 Å) | | Cite: | Structural insights into Parkin substrate lysine targeting from minimal Miro substrates.
Sci Rep, 6, 2016
|
|
5KNM
 
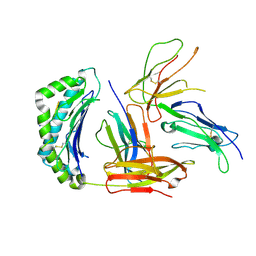 | |
3HE0
 
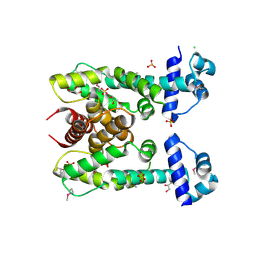 | | The Structure of a Putative Transcriptional Regulator TetR Family Protein from Vibrio parahaemolyticus. | | Descriptor: | CHLORIDE ION, GLYCEROL, SULFATE ION, ... | | Authors: | Cuff, M.E, Hendricks, R, Moy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-05-07 | | Release date: | 2009-07-07 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Structure of a Putative Transcriptional Regulator TetR Family Protein from Vibrio parahaemolyticus.
TO BE PUBLISHED
|
|
5KO0
 
 | | Human Islet Amyloid Polypeptide Segment 15-FLVHSSNNFGA-25 Determined by MicroED | | Descriptor: | THIOCYANATE ION, hIAPP(15-25)WT | | Authors: | Krotee, P.A.L, Rodriguez, J.A, Sawaya, M.R, Cascio, D, Shi, D, Nannenga, B.L, Hattne, J, Reyes, F.E, Gonen, T, Eisenberg, D.S. | | Deposit date: | 2016-06-28 | | Release date: | 2016-12-21 | | Last modified: | 2024-03-06 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.4 Å) | | Cite: | Atomic structures of fibrillar segments of hIAPP suggest tightly mated beta-sheets are important for cytotoxicity.
Elife, 6, 2017
|
|
3HK5
 
 | | Crystal structure of uronate isomerase from Bacillus halodurans complexed with zinc and D-Arabinarate | | Descriptor: | CARBONATE ION, CHLORIDE ION, D-arabinaric acid, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Nguyen, T.T, Raushel, F.M, Almo, S.C. | | Deposit date: | 2009-05-22 | | Release date: | 2009-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The mechanism of the reaction catalyzed by uronate isomerase illustrates how an isomerase may have evolved from a hydrolase within the amidohydrolase superfamily.
Biochemistry, 48, 2009
|
|
5KRS
 
 | | HIV-1 Integrase Catalytic Core Domain in Complex with an Allosteric Inhibitor, 3-(1H-pyrrol-1-yl)-2-thiophenecarboxylic acid | | Descriptor: | 3-pyrrol-1-ylthiophene-2-carboxylic acid, DIMETHYL SULFOXIDE, Integrase | | Authors: | Patel, D, Bauman, J.D, Arnold, E. | | Deposit date: | 2016-07-07 | | Release date: | 2016-09-28 | | Last modified: | 2022-03-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A New Class of Allosteric HIV-1 Integrase Inhibitors Identified by Crystallographic Fragment Screening of the Catalytic Core Domain.
J.Biol.Chem., 291, 2016
|
|
3HK8
 
 | | Crystal structure of uronate isomerase from Bacillus halodurans complexed with zinc and D-Arabinohydroxamate | | Descriptor: | CARBONATE ION, CHLORIDE ION, D-arabinohydroxamic acid, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Nguyen, T.T, Raushel, F.M, Almo, S.C. | | Deposit date: | 2009-05-22 | | Release date: | 2009-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The mechanism of the reaction catalyzed by uronate isomerase illustrates how an isomerase may have evolved from a hydrolase within the amidohydrolase superfamily.
Biochemistry, 48, 2009
|
|
