5HJN
 
 | |
5KTG
 
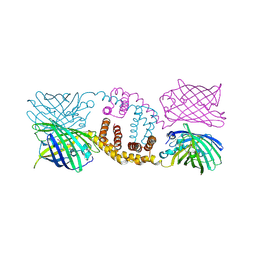 | |
5BT0
 
 | | Switching GFP fluorescence using genetically encoded phenyl azide chemistry through two different non-native post-translational modifications routes at the same position. | | Descriptor: | Green fluorescent protein, SULFATE ION | | Authors: | Hartley, A.M, Worthy, H.L, Reddington, S.C, Rizkallah, P.J, Jones, D.D. | | Deposit date: | 2015-06-02 | | Release date: | 2016-07-13 | | Last modified: | 2017-05-10 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Molecular basis for functional switching of GFP by two disparate non-native post-translational modifications of a phenyl azide reaction handle.
Chem Sci, 7, 2016
|
|
5DY6
 
 | | Enhanced superfolder GFP with DBCO at 148 | | Descriptor: | Green fluorescent protein, SULFATE ION | | Authors: | Jones, D.D, Rizkallah, P.J, Worthy, H.L. | | Deposit date: | 2015-09-24 | | Release date: | 2016-07-13 | | Last modified: | 2018-05-30 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Molecular basis for functional switching of GFP by two disparate non-native post-translational modifications of a phenyl azide reaction handle.
Chem Sci, 7, 2016
|
|
5BTT
 
 | | Switching GFP fluorescence using genetically encoded phenyl azide chemistry through two different non-native post-translational modifications routes at the same position. | | Descriptor: | GLYCEROL, Green fluorescent protein, SULFATE ION | | Authors: | Hartley, A.M, Worthy, H.L, Reddington, S.C, Rizkallah, P.J, Jones, D.D. | | Deposit date: | 2015-06-03 | | Release date: | 2016-07-13 | | Last modified: | 2017-05-10 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Molecular basis for functional switching of GFP by two disparate non-native post-translational modifications of a phenyl azide reaction handle.
Chem Sci, 7, 2016
|
|
5FC8
 
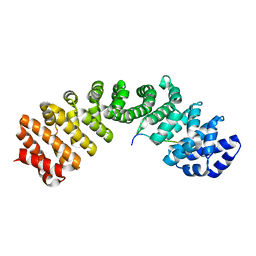 | | Mouse importin alpha: Dengue 3 NS5 C-terminal NLS peptide complex | | Descriptor: | Importin subunit alpha-1, Nonstructural protein 5 | | Authors: | Smith, K.M, Forwood, J.K. | | Deposit date: | 2015-12-15 | | Release date: | 2016-05-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The C-terminal 18 Amino Acid Region of Dengue Virus NS5 Regulates its Subcellular Localization and Contains a Conserved Arginine Residue Essential for Infectious Virus Production.
PLoS Pathog., 12, 2016
|
|
5HHG
 
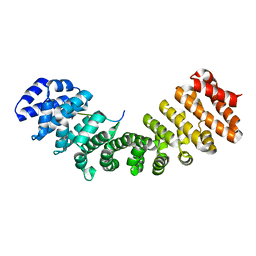 | | Mouse importin alpha: Dengue 2 NS5 C-terminal NLS peptide complex | | Descriptor: | Importin subunit alpha-1, RNA-directed RNA polymerase NS5 | | Authors: | Smith, K.M, Forwood, J.K. | | Deposit date: | 2016-01-11 | | Release date: | 2016-05-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The C-terminal 18 Amino Acid Region of Dengue Virus NS5 Regulates its Subcellular Localization and Contains a Conserved Arginine Residue Essential for Infectious Virus Production.
PLoS Pathog., 12, 2016
|
|
5JG4
 
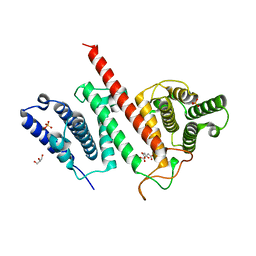 | | Structure of the effector protein LpiR1 (Lpg0634) from Legionella pneumophila | | Descriptor: | CITRATE ANION, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Beyrakhova, K, van Straaten, K, Cygler, M. | | Deposit date: | 2016-04-19 | | Release date: | 2016-05-04 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and Functional Investigations of the Effector Protein LpiR1 from Legionella pneumophila.
J.Biol.Chem., 291, 2016
|
|
4ZVR
 
 | |
4ZVT
 
 | |
4ZVS
 
 | |
4ZVP
 
 | |
4ZVO
 
 | |
4ZVQ
 
 | |
4ZVU
 
 | |
5IH4
 
 | | Human Casein Kinase 1 isoform delta apo (kinase domain) | | Descriptor: | Casein kinase I isoform delta, S,R MESO-TARTARIC ACID, SULFATE ION, ... | | Authors: | Ursu, A, Illich, D.J, Takemoto, Y, Porfetye, A.T, Zhang, M, Brockmeyer, A, Janning, P, Watanabe, N, Osada, H, Vetter, I.R, Ziegler, S, Schoeler, H.R, Waldmann, H. | | Deposit date: | 2016-02-29 | | Release date: | 2016-04-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Epiblastin A Induces Reprogramming of Epiblast Stem Cells Into Embryonic Stem Cells by Inhibition of Casein Kinase 1.
Cell Chem Biol, 23, 2016
|
|
5IH5
 
 | | Human Casein Kinase 1 isoform delta (kinase domain) in complex with Epiblastin A | | Descriptor: | 6-(3-chlorophenyl)pteridine-2,4,7-triamine, Casein kinase I isoform delta, S,R MESO-TARTARIC ACID, ... | | Authors: | Ursu, A, Illich, D.J, Takemoto, Y, Porfetye, A.T, Zhang, M, Brockmeyer, A, Janning, P, Watanabe, N, Osada, H, Vetter, I.R, Ziegler, S, Schoeler, H.R, Waldmann, H. | | Deposit date: | 2016-02-29 | | Release date: | 2016-04-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Epiblastin A Induces Reprogramming of Epiblast Stem Cells Into Embryonic Stem Cells by Inhibition of Casein Kinase 1.
Cell Chem Biol, 23, 2016
|
|
5IH6
 
 | | Human Casein Kinase 1 isoform delta (kinase domain) in complex with Epiblastin A derivative | | Descriptor: | 6-(3-bromophenyl)pteridine-2,4,7-triamine, Casein kinase I isoform delta, S,R MESO-TARTARIC ACID, ... | | Authors: | Ursu, A, Illich, D.J, Takemoto, Y, Porfetye, A.T, Zhang, M, Brockmeyer, A, Janning, P, Watanabe, N, Osada, H, Vetter, I.R, Ziegler, S, Schoeler, H.R, Waldmann, H. | | Deposit date: | 2016-02-29 | | Release date: | 2016-04-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Epiblastin A Induces Reprogramming of Epiblast Stem Cells Into Embryonic Stem Cells by Inhibition of Casein Kinase 1.
Cell Chem Biol, 23, 2016
|
|
5J2O
 
 | |
5FVI
 
 | | Structure of IrisFP in mineral grease at 100 K. | | Descriptor: | Green to red photoconvertible GFP-like protein EosFP, SULFATE ION | | Authors: | Colletier, J.P, Gallat, F.X, Coquelle, N, Weik, M. | | Deposit date: | 2016-02-07 | | Release date: | 2016-04-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.397 Å) | | Cite: | Serial Femtosecond Crystallography and Ultrafast Absorption Spectroscopy of the Photoswitchable Fluorescent Protein Irisfp.
J.Phys.Chem.Lett., 7, 2016
|
|
5FVF
 
 | | Room temperature structure of IrisFP determined by serial femtosecond crystallography. | | Descriptor: | AMMONIUM ION, Green to red photoconvertible GFP-like protein EosFP, SULFATE ION | | Authors: | Colletier, J.P, Gallat, F.X, Coquelle, N, Weik, M. | | Deposit date: | 2016-02-06 | | Release date: | 2016-04-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Serial Femtosecond Crystallography and Ultrafast Absorption Spectroscopy of the Photoswitchable Fluorescent Protein Irisfp.
J.Phys.Chem.Lett., 7, 2016
|
|
4Z4K
 
 | |
5EB6
 
 | |
5EJU
 
 | |
5EB7
 
 | |
