4PC1
 
 | | Elongation Factor Tu:Ts complex with a bound phosphate | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Elongation factor Ts, Elongation factor Tu, ... | | Authors: | Thirup, S.S. | | Deposit date: | 2014-04-14 | | Release date: | 2015-05-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural outline of the detailed mechanism for elongation factor Ts-mediated guanine nucleotide exchange on elongation factor Tu.
J.Struct.Biol., 191, 2015
|
|
5AK7
 
 | | Structure of wt Porphyromonas gingivalis peptidylarginine deiminase | | Descriptor: | 1,2-ETHANEDIOL, ALANINE, ARGININE, ... | | Authors: | Kopec, J, Montgomery, A, Shrestha, L, Kiyani, W, Nowak, R, Burgess-Brown, N, Venables, P.J, Yue, W.W. | | Deposit date: | 2015-03-02 | | Release date: | 2015-07-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Crystal Structure of Porphyromonas Gingivalis Peptidylarginine Deiminase: Implications for Autoimmunity in Rheumatoid Arthritis.
Ann.Rheum.Dis., 75, 2016
|
|
7FR1
 
 | |
7FRB
 
 | |
5SOH
 
 | |
5SNK
 
 | |
4ZIY
 
 | |
9J76
 
 | |
4BQ3
 
 | | Structural analysis of an exo-beta-agarase | | Descriptor: | B-AGARASE, CALCIUM ION, GLYCEROL, ... | | Authors: | Pluvinage, B, Hehemann, J.H, Boraston, A.B. | | Deposit date: | 2013-05-29 | | Release date: | 2013-08-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Substrate Recognition and Hydrolysis by a Family 50 Exo-Beta-Agarase Aga50D from the Marine Bacterium Saccharophagus Degradans
J.Biol.Chem., 288, 2013
|
|
2VAX
 
 | | Crystal structure of deacetylcephalosporin C acetyltransferase (Cephalosporin C-soak) | | Descriptor: | 4-(3-ACETOXYMETHYL-2-CARBOXY-8-OXO-5-THIA-1-AZA-BICYCLO[4.2.0]OCT-2-EN-7-YLCARBAMOYL)-1-CARBOXY-BUTYL-AMMONIUM, ACETATE ION, ACETYL-COA--DEACETYLCEPHALOSPORIN C ACETYLTRANSFERASE | | Authors: | Lejon, S, Ellis, J, Valegard, K. | | Deposit date: | 2007-09-04 | | Release date: | 2008-09-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The Last Step in Cephalosporin C Formation Revealed: Crystal Structures of Deacetylcephalosporin C Acetyltransferase from Acremonium Chrysogenum in Complexes with Reaction Intermediates.
J.Mol.Biol., 377, 2008
|
|
5SWP
 
 | | Crystal Structure of PI3Kalpha in complex with fragments 6 and 24 | | Descriptor: | 2-methylcyclohexane-1,3-dione, CHLORIDE ION, Phosphatidylinositol 3-kinase regulatory subunit alpha, ... | | Authors: | Gabelli, S.B, Vogelstein, B, Miller, M.S, Amzel, L.M. | | Deposit date: | 2016-08-08 | | Release date: | 2017-02-15 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.41 Å) | | Cite: | Identification of allosteric binding sites for PI3K alpha oncogenic mutant specific inhibitor design.
Bioorg. Med. Chem., 25, 2017
|
|
1ANR
 
 | |
3IXS
 
 | | Ring1B C-terminal domain/RYBP C-terminal domain Complex | | Descriptor: | 1,2-ETHANEDIOL, 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, E3 ubiquitin-protein ligase RING2, ... | | Authors: | Wang, R, Taylor, A.B, Kim, C.A. | | Deposit date: | 2009-09-04 | | Release date: | 2010-08-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Polycomb Group Targeting through Different Binding Partners of RING1B C-Terminal Domain.
Structure, 18, 2010
|
|
5T28
 
 | | mPI3Kd IN COMPLEX WITH 5k | | Descriptor: | 3-(benzotriazol-1-yl)-5-[1-methyl-5-(1-phenylcyclopropyl)-1,2,4-triazol-3-yl]pyrazin-2-amine, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform | | Authors: | Petersen, J, Terstige, I, Perry, M, Svensson, T, Tyrchan, C, Lindmark, H, Oster, L. | | Deposit date: | 2016-08-23 | | Release date: | 2017-04-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery of triazole aminopyrazines as a highly potent and selective series of PI3K delta inhibitors.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
4Z3V
 
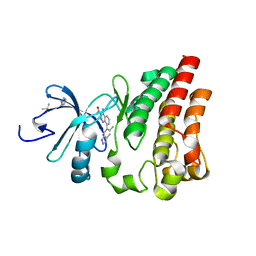 | |
4P9J
 
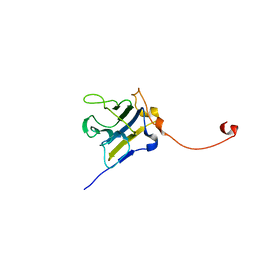 | |
4BSN
 
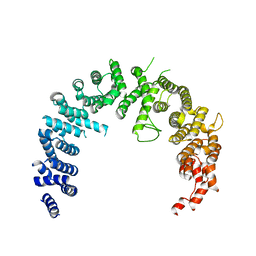 | | Crystal structure of the Nuclear Export Receptor CRM1 (exportin-1) lacking the C-terminal helical extension at 4.1A | | Descriptor: | EXPORTIN-1 | | Authors: | Dian, C, Bernaudat, F, Langer, K, Oliva, M.F, Fornerod, M, Schoehn, G, Muller, C.W, Petosa, C. | | Deposit date: | 2013-06-11 | | Release date: | 2013-07-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (4.1 Å) | | Cite: | Structure of a Truncation Mutant of the Nuclear Export Factor Crm1 Provides Insights Into the Auto-Inhibitory Role of its C-Terminal Helix.
Structure, 21, 2013
|
|
5WAE
 
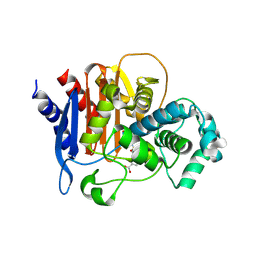 | | ADC-7 in complex with boronic acid transition state inhibitor CR167 | | Descriptor: | 3-(5,7,7-trihydroxy-2,2,7-trioxo-6-oxa-2lambda~6~-thia-3-aza-7lambda~5~-phospha-5-boraheptan-1-yl)benzoic acid, Beta-lactamase | | Authors: | Powers, R.A, Wallar, B.J. | | Deposit date: | 2017-06-26 | | Release date: | 2017-12-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.804 Å) | | Cite: | Structure-Based Analysis of Boronic Acids as Inhibitors of Acinetobacter-Derived Cephalosporinase-7, a Unique Class C beta-Lactamase.
ACS Infect Dis, 4, 2018
|
|
2UZ8
 
 | | The crystal structure of p18, human translation elongation factor 1 epsilon 1 | | Descriptor: | EUKARYOTIC TRANSLATION ELONGATION FACTOR 1 EPSILON-1, GLYCEROL | | Authors: | Kang, B.S, Kim, K.J, Kim, M.H, Oh, Y.S, Kim, S. | | Deposit date: | 2007-04-26 | | Release date: | 2008-03-25 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Determination of Three-Dimensional Structure and Residues of the Novel Tumor Suppressor Aimp3/P18 Required for the Interaction with Atm.
J.Biol.Chem., 283, 2008
|
|
1NMA
 
 | | N9 NEURAMINIDASE COMPLEXES WITH ANTIBODIES NC41 AND NC10: EMPIRICAL FREE-ENERGY CALCULATIONS CAPTURE SPECIFICITY TRENDS OBSERVED WITH MUTANT BINDING DATA | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, FAB NC10, N9 NEURAMINIDASE, ... | | Authors: | Tulip, W.R, Varghese, J.N, Colman, P.M. | | Deposit date: | 1994-05-06 | | Release date: | 1995-09-15 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | N9 neuraminidase complexes with antibodies NC41 and NC10: empirical free energy calculations capture specificity trends observed with mutant binding data.
Biochemistry, 33, 1994
|
|
6UFU
 
 | | C2 symmetric peptide design number 1, Zappy, crystal form 1 | | Descriptor: | C2-1, Zappy, crystal form 1 | | Authors: | Mulligan, V.K, Kang, C.S, Antselovich, I, Sawaya, M.R, Yeates, T.O, Baker, D. | | Deposit date: | 2019-09-25 | | Release date: | 2020-12-02 | | Last modified: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (1.101 Å) | | Cite: | Computational design of mixed chirality peptide macrocycles with internal symmetry.
Protein Sci., 29, 2020
|
|
5SZX
 
 | | Epstein-Barr virus Zta DNA binding domain homodimer in complex with methylated DNA | | Descriptor: | DNA (5'-D(*AP*AP*GP*CP*AP*CP*TP*GP*AP*GP*(5CM)P*GP*AP*TP*GP*AP*AP*G)-3'), DNA (5'-D(*TP*CP*TP*TP*CP*AP*TP*(5CM)P*GP*CP*TP*CP*AP*GP*TP*GP*CP*T)-3'), PHOSPHATE ION, ... | | Authors: | Hong, S, Horton, J.R, Cheng, X. | | Deposit date: | 2016-08-15 | | Release date: | 2017-03-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.251 Å) | | Cite: | Methyl-dependent and spatial-specific DNA recognition by the orthologous transcription factors human AP-1 and Epstein-Barr virus Zta.
Nucleic Acids Res., 45, 2017
|
|
2B5G
 
 | | Wild Type SSAT- 1.7A structure | | Descriptor: | Diamine acetyltransferase 1, SULFATE ION | | Authors: | Bewley, M.C, Graziano, V, Jiang, J.S, Matz, E, Studier, F.W, Pegg, A.P, Coleman, C.S, Flanagan, J.M, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2005-09-28 | | Release date: | 2006-01-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structures of wild-type and mutant human spermidine/spermine N1-acetyltransferase, a potential therapeutic drug target
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
5CTM
 
 | | Structure of BPu1 beta-lactamase | | Descriptor: | 1,2-ETHANEDIOL, Beta-lactamase, CITRATE ANION, ... | | Authors: | Smith, C.A, Vakulenko, S.B. | | Deposit date: | 2015-07-24 | | Release date: | 2015-11-18 | | Last modified: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Class D beta-lactamases do exist in Gram-positive bacteria.
Nat.Chem.Biol., 12, 2016
|
|
5CY8
 
 | | Structure of Xoo1075, a peptide deformylase from Xanthomonas oryzae pv oryze, in complex with fragment 244 | | Descriptor: | (3R)-2,3-dihydro[1,3]thiazolo[3,2-a]benzimidazol-3-ol, ACETATE ION, CADMIUM ION, ... | | Authors: | Ngo, H.P.T, Kang, L.W. | | Deposit date: | 2015-07-30 | | Release date: | 2016-08-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Structure of Xoo1075, a peptide deformylase from Xanthomonas oryzae pv oryze, in complex with fragment 244
To Be Published
|
|
