2XKX
 
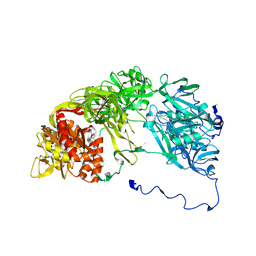 | | Single particle analysis of PSD-95 in negative stain | | Descriptor: | DISKS LARGE HOMOLOG 4 | | Authors: | Fomina, S, Howard, T.D, Sleator, O.K, Golovanova, M, O'Ryan, L, Leyland, M.L, Grossmann, J.G, Collins, R.F, Prince, S.M. | | Deposit date: | 2010-07-15 | | Release date: | 2011-07-20 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (22.9 Å), SOLUTION SCATTERING | | Cite: | Self-Directed Assembly and Clustering of the Cytoplasmic Domains of Inwardly Rectifying Kir2.1 Potassium Channels on Association with Psd-95
Biochim.Biophys.Acta, 1808, 2011
|
|
6KCT
 
 | |
4CDX
 
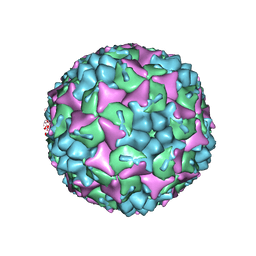 | | Crystal structure of human Enterovirus 71 in complex with the uncoating inhibitor GPP12 | | Descriptor: | 1-(5-((3'-METHYL-[1,1'-BIPHENYL]-4-YL)OXY)PENTYL)-3-(, SODIUM ION, VP1, ... | | Authors: | De Colibus, L, Wang, X, Spyrou, J.A.B, Kelly, J, Ren, J, Grimes, J, Puerstinger, G, Stonehouse, N, Walter, T.S, Hu, Z, Wang, J, Li, X, Peng, W, Rowlands, D, Fry, E.E, Rao, Z, Stuart, D.I. | | Deposit date: | 2013-11-07 | | Release date: | 2014-02-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | More-Powerful Virus Inhibitors from Structure-Based Analysis of Hev71 Capsid-Binding Molecules.
Nat.Struct.Mol.Biol., 21, 2014
|
|
1ZJA
 
 | | Crystal structure of the trehalulose synthase MutB from Pseudomonas mesoacidophila MX-45 (triclinic form) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, Trehalulose synthase | | Authors: | Ravaud, S, Robert, X, Haser, R, Aghajari, N. | | Deposit date: | 2005-04-28 | | Release date: | 2006-10-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Trehalulose synthase native and carbohydrate complexed structures provide insights into sucrose isomerization.
J.Biol.Chem., 282, 2007
|
|
5BSV
 
 | |
4PPS
 
 | | Crystal Structure of the Estrogen Receptor alpha Ligand-binding Domain in Complex with an A-CD ring estrogen derivative | | Descriptor: | (1S,3aR,5R,7aS)-5-(4-hydroxyphenyl)-7a-methyloctahydro-1H-inden-1-ol, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Nwachukwu, J.C, Srinivasan, S, Bruno, N.E, Parent, A.A, Hughes, T.S, Pollock, J.A, Gjyshi, O, Cavett, V, Nowak, J, Garcia-Ordonez, R.D, Houtman, R, Griffin, P.R, Kojetin, D.J, Katzenellenbogen, J.A, Conkright, M.D, Nettles, K.W. | | Deposit date: | 2014-02-27 | | Release date: | 2014-05-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.929 Å) | | Cite: | Resveratrol modulates the inflammatory response via an estrogen receptor-signal integration network.
Elife, 3, 2014
|
|
2XGB
 
 | | Crystal structure of Barley Beta-Amylase complexed with 2,3- epoxypropyl-alpha-D-glucopyranoside | | Descriptor: | (2R)-oxiran-2-ylmethyl alpha-D-glucopyranoside, 1,2-ETHANEDIOL, BETA-AMYLASE | | Authors: | Rejzek, M, Stevenson, C.E.M, Southard, A.M, Stanley, D, Denyer, K, Smith, A.M, Naldrett, M.J, Lawson, D.M, Field, R.A. | | Deposit date: | 2010-06-02 | | Release date: | 2010-12-01 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Chemical Genetics and Cereal Starch Metabolism: Structural Basis of the Non-Covalent and Covalent Inhibition of Barley Beta-Amylase.
Mol.Biosyst., 7, 2011
|
|
2XGI
 
 | | Crystal structure of Barley Beta-Amylase complexed with 3,4- epoxybutyl alpha-D-glucopyranoside | | Descriptor: | (3R)-3-hydroxybutyl alpha-D-glucopyranoside, (3S)-3-hydroxybutyl alpha-D-glucopyranoside, 1,2-ETHANEDIOL, ... | | Authors: | Rejzek, M, Stevenson, C.E.M, Southard, A.M, Stanley, D, Denyer, K, Smith, A.M, Naldrett, M.J, Lawson, D.M, Field, R.A. | | Deposit date: | 2010-06-04 | | Release date: | 2010-12-01 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Chemical genetics and cereal starch metabolism: structural basis of the non-covalent and covalent inhibition of barley beta-amylase.
Mol Biosyst, 7, 2011
|
|
4POK
 
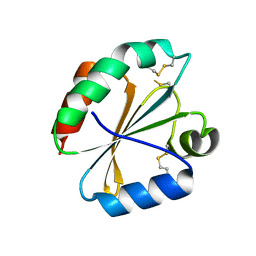 | | Crystal structures of thioredoxin with mesna at 2.5A resolution | | Descriptor: | 1-THIOETHANESULFONIC ACID, Thioredoxin | | Authors: | Sridhar, V, Chie-Leon, B, Badger, J, Nienaber, V.L, Hausheer, F.H. | | Deposit date: | 2014-02-25 | | Release date: | 2014-10-29 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | BNP7787 Forms Novel Covalent
Adducts on Human Thioredoxin
and Modulates Thioredoxin
Activity
J Pharmacol Clin Toxicol, 2, 2014
|
|
6KI9
 
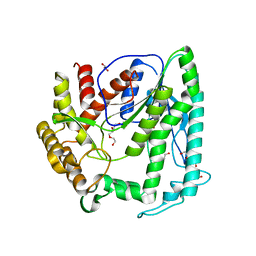 | | Apo structure of FabMG, novel types of Enoyl-acyl carrier protein reductase | | Descriptor: | 1,2-ETHANEDIOL, FabMG, novel types of Enoyl-acyl carrier protein reductase, ... | | Authors: | Kim, S, Rhee, S. | | Deposit date: | 2019-07-17 | | Release date: | 2020-05-20 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | A triclosan-resistance protein from the soil metagenome is a novel enoyl-acyl carrier protein reductase: Structure-guided functional analysis.
Febs J., 287, 2020
|
|
4PPB
 
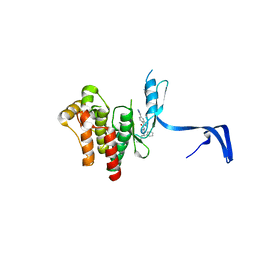 | |
6KFZ
 
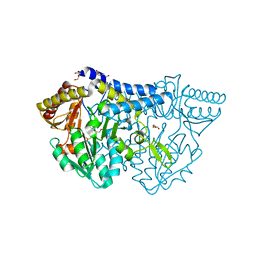 | | SufS from Bacillus subtilis, soaked with L-cysteine for 90 sec at 1.96 angstrom resolution | | Descriptor: | Cysteine desulfurase SufS, DI(HYDROXYETHYL)ETHER, N-({3-HYDROXY-2-METHYL-5-[(PHOSPHONOOXY)METHYL]PYRIDIN-4-YL}METHYL)-L-CYSTEINE | | Authors: | Nakamura, R, Takahashi, Y, Fujishiro, T. | | Deposit date: | 2019-07-09 | | Release date: | 2019-10-16 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Snapshots of PLP-substrate and PLP-product external aldimines as intermediates in two types of cysteine desulfurase enzymes.
Febs J., 287, 2020
|
|
4XI8
 
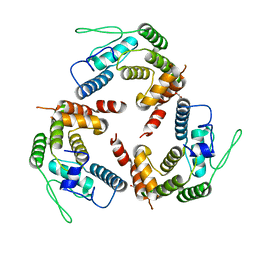 | |
5BJW
 
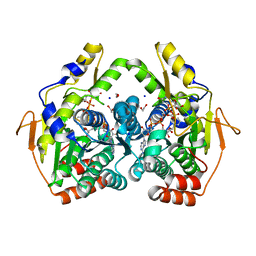 | | X-ray structure of the PglF 4,6-dehydratase from campylobacter jejuni, T595S variant, in complex with UDP | | Descriptor: | 1,2-ETHANEDIOL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SODIUM ION, ... | | Authors: | Riegert, A.S, Thoden, J.B, Holden, H.M. | | Deposit date: | 2017-09-12 | | Release date: | 2017-11-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and Biochemical Investigation of PglF from Campylobacter jejuni Reveals a New Mechanism for a Member of the Short Chain Dehydrogenase/Reductase Superfamily.
Biochemistry, 56, 2017
|
|
4PS6
 
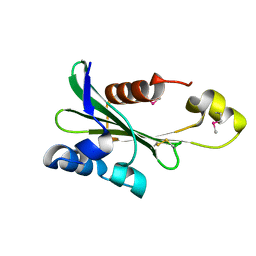 | |
1ZOP
 
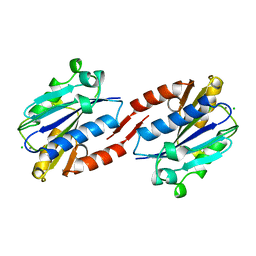 | | CD11A I-DOMAIN WITH BOUND MAGNESIUM ION | | Descriptor: | CHLORIDE ION, I-DOMAIN FRAGMENT OF LFA-1, MANGANESE (II) ION | | Authors: | Leahy, D.J, Qu, A. | | Deposit date: | 1996-06-21 | | Release date: | 1996-12-07 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The role of the divalent cation in the structure of the I domain from the CD11a/CD18 integrin.
Structure, 4, 1996
|
|
4XDQ
 
 | |
2XNK
 
 | |
5XU2
 
 | | Crystal Structure of Transketolase in complex with TPP_III and fructose-6-phosphate from Pichia Stipitis | | Descriptor: | 2-[3-[(4-azanyl-2-methyl-pyrimidin-5-yl)methyl]-4-methyl-2H-1,3-thiazol-5-yl]ethyl phosphono hydrogen phosphate, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Li, T.L, Hsu, N.S, Wang, Y.L. | | Deposit date: | 2017-06-22 | | Release date: | 2018-06-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | Evidence of Diradicals Involved in the Yeast Transketolase Catalyzed Keto-Transferring Reactions.
Chembiochem, 19, 2018
|
|
4XEW
 
 | | Crystal structure of 7,8-diaminopelargonic acid synthase (BioA) from Mycobacterium tuberculosis, complexed with a HTS lead compound | | Descriptor: | 6-(2-fluorophenyl)[1,3]dioxolo[4,5-g]quinolin-8(5H)-one, Adenosylmethionine-8-amino-7-oxononanoate aminotransferase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Finzel, B.C, Dai, R. | | Deposit date: | 2014-12-25 | | Release date: | 2015-07-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Fragment-Based Exploration of Binding Site Flexibility in Mycobacterium tuberculosis BioA.
J.Med.Chem., 58, 2015
|
|
7EG1
 
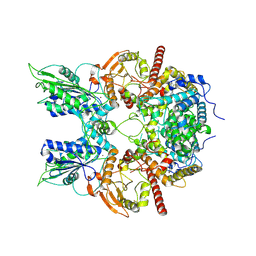 | | Cryo-EM structure of DNMDP-induced PDE3A-SLFN12 complex | | Descriptor: | (4~{R})-3-[4-(diethylamino)-3-[oxidanyl(oxidanylidene)-$l^{4}-azanyl]phenyl]-4-methyl-4,5-dihydro-1~{H}-pyridazin-6-one, MAGNESIUM ION, Schlafen family member 12, ... | | Authors: | Liu, N, Chen, J, Wang, X.D, Wang, H.W. | | Deposit date: | 2021-03-23 | | Release date: | 2021-11-03 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure of PDE3A-SLFN12 complex and structure-based design for a potent apoptosis inducer of tumor cells.
Nat Commun, 12, 2021
|
|
170L
 
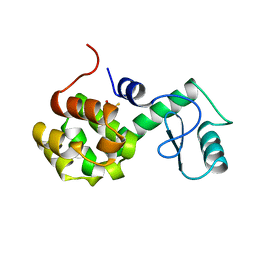 | | PROTEIN FLEXIBILITY AND ADAPTABILITY SEEN IN 25 CRYSTAL FORMS OF T4 LYSOZYME | | Descriptor: | BETA-MERCAPTOETHANOL, T4 LYSOZYME | | Authors: | Zhang, X.-J, Weaver, L.H, Wozniak, A, Matthews, B.W. | | Deposit date: | 1995-03-24 | | Release date: | 1995-07-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Protein flexibility and adaptability seen in 25 crystal forms of T4 lysozyme.
J.Mol.Biol., 250, 1995
|
|
3HN5
 
 | |
6DQP
 
 | |
4Q5J
 
 | | Crystal structure of SeMet derivative BRI1 in complex with BKI1 | | Descriptor: | BRI1 kinase inhibitor 1, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Protein BRASSINOSTEROID INSENSITIVE 1 | | Authors: | Wang, J, Wang, J, Chen, L, Wu, J.W, Wang, Z.X. | | Deposit date: | 2014-04-17 | | Release date: | 2014-10-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.772 Å) | | Cite: | Structural insights into the negative regulation of BRI1 signaling by BRI1-interacting protein BKI1.
Cell Res., 24, 2014
|
|
