7ADF
 
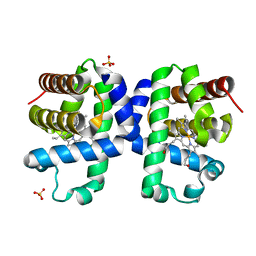 | | SFX structure of dehaloperoxidase B in the ferric form | | Descriptor: | Dehaloperoxidase B, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Moreno Chicano, T, Ebrahim, A, Axford, D.A, Sherrell, D.A, Sugimoto, H, Tono, K, Owada, S, Worrall, J.W, Strange, R.W, Owen, R.L, Hough, M.A. | | Deposit date: | 2020-09-14 | | Release date: | 2021-10-06 | | Last modified: | 2025-01-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | SFX structure of dehaloperoxidase B in the ferric form
to be published
|
|
2BEU
 
 | | Reactivity modulation of human branched-chain alpha-ketoacid dehydrogenase by an internal molecular switch | | Descriptor: | 2-OXOISOVALERATE DEHYDROGENASE ALPHA SUBUNIT, 2-OXOISOVALERATE DEHYDROGENASE BETA SUBUNIT, C2-1-HYDROXY-3-METHYL-PROPYL-THIAMIN DIPHOSPHATE, ... | | Authors: | Machius, M, Wynn, R.M, Chuang, J.L, Tomchick, D.R, Brautigam, C.A, Chuang, D.T. | | Deposit date: | 2004-11-30 | | Release date: | 2006-02-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | A Versatile Conformational Switch Regulates Reactivity in Human Branched-Chain Alpha-Ketoacid Dehydrogenase.
Structure, 14, 2006
|
|
2BTR
 
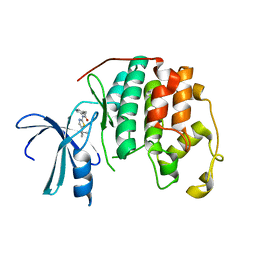 | | STRUCTURE OF CDK2 COMPLEXED WITH PNU-198873 | | Descriptor: | CELL DIVISION PROTEIN KINASE 2, N-(5-ISOPROPYL-THIAZOL-2-YL)-2-PYRIDIN-3-YL-ACETAMIDE | | Authors: | Vulpetti, A, Casale, E, Roletto, F, Amici, R, Villa, M, Pevarello, P. | | Deposit date: | 2005-06-06 | | Release date: | 2005-11-09 | | Last modified: | 2025-09-10 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure-Based Drug Design to the Discovery of New 2-Aminothiazole Cdk2 Inhibitors.
J.Mol.Graph.Model., 24, 2006
|
|
6D1E
 
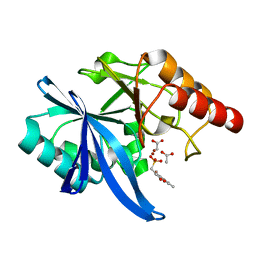 | | Crystal structure of NDM-1 complexed with compound 7 | | Descriptor: | ACETATE ION, Metallo-beta-lactamase type 2, ZINC ION, ... | | Authors: | Pemberton, O.A, Chen, Y. | | Deposit date: | 2018-04-11 | | Release date: | 2019-04-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Heteroaryl Phosphonates as Noncovalent Inhibitors of Both Serine- and Metallocarbapenemases.
J.Med.Chem., 62, 2019
|
|
6D1K
 
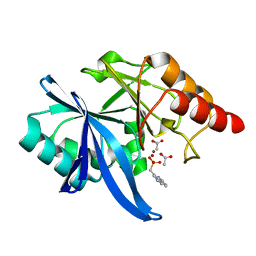 | |
2BGK
 
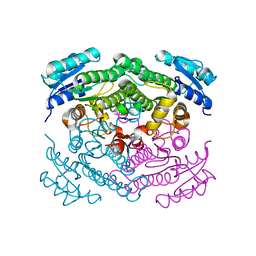 | | X-Ray structure of apo-Secoisolariciresinol Dehydrogenase | | Descriptor: | RHIZOME SECOISOLARICIRESINOL DEHYDROGENASE | | Authors: | Youn, B, Moinuddin, S.G, Davin, L.B, Lewis, N.G, Kang, C. | | Deposit date: | 2004-12-23 | | Release date: | 2005-01-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structures of Apo-Form and Binary/Ternary Complexes of Podophyllum Secoisolariciresinol Dehydrogenase, an Enzyme Involved in Formation of Health-Protecting and Plant Defense Lignans
J.Biol.Chem., 280, 2005
|
|
7A8Q
 
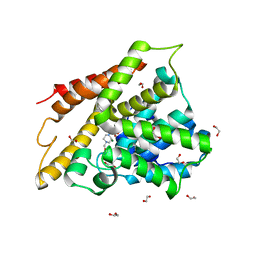 | | Crystal structure of human phosphodiesterase 4D2 catalytic domain with inhibitor NPD-654 | | Descriptor: | 1,2-ETHANEDIOL, 2-cycloheptyl-5-[4-methoxy-3-[[4-(1~{H}-1,2,3,4-tetrazol-5-yl)phenyl]methoxy]phenyl]-4,4-dimethyl-pyrazolidin-3-one, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Singh, A.K, Blaazer, A.R, Zara, L, de Esch, I.J.P, Leurs, R, Brown, D.G. | | Deposit date: | 2020-08-30 | | Release date: | 2021-10-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | hPDE4D2 structure with inhibitor NPD-654
To be published
|
|
6CTZ
 
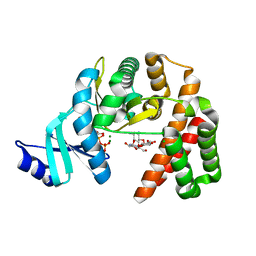 | | Structure of the GDP and kanamycin complex of APH(2")-IIia | | Descriptor: | CHLORIDE ION, GUANOSINE-5'-DIPHOSPHATE, Gentamicin resistance protein, ... | | Authors: | Smith, C.A, Vakulenko, S.B. | | Deposit date: | 2018-03-23 | | Release date: | 2019-03-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Structural basis for the diversity of the mechanism of nucleotide hydrolysis by the aminoglycoside-2''-phosphotransferases
Acta Crystallogr.,Sect.D, 75, 2019
|
|
2BGT
 
 | |
2C29
 
 | | Structure of dihydroflavonol reductase from Vitis vinifera at 1.8 A. | | Descriptor: | (2R,3R)-2-(3,4-DIHYDROXYPHENYL)-3,5,7-TRIHYDROXY-2,3-DIHYDRO-4H-CHROMEN-4-ONE, DIHYDROFLAVONOL 4-REDUCTASE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Petit, P, Granier, T, D'Estaintot, B.L, Hamdi, S, Gallois, B. | | Deposit date: | 2005-09-27 | | Release date: | 2006-10-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Crystal Structure of Grape Dihydroflavonol 4-Reductase, a Key Enzyme in Flavonoid Biosynthesis.
J.Mol.Biol., 368, 2007
|
|
7A9V
 
 | | Crystal structure of human phosphodiesterase 4D2 catalytic domain with inhibitor NPD-635 | | Descriptor: | 1,2-ETHANEDIOL, 1-cycloheptyl-3-[4-methoxy-3-(2-phenylethoxy)phenyl]-4,4-dimethyl-4,5-dihydro-1H-pyrazol-5-one, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Singh, A.K, Blaazer, A.R, Zara, L, de Esch, I.J.P, Leurs, R, Brown, D.G. | | Deposit date: | 2020-09-02 | | Release date: | 2021-10-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | hPDE4D2 structure with inhibitor NPD-635
To be published
|
|
2BLL
 
 | | Apo-structure of the C-terminal decarboxylase domain of ArnA | | Descriptor: | PROTEIN YFBG | | Authors: | Williams, G.J, Breazeale, S.D, Raetz, C.R.H, Naismith, J.H. | | Deposit date: | 2005-03-07 | | Release date: | 2005-04-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and Function of Both Domains of Arna, a Dual Function Decarboxylase and a Formyltransferase, Involved in 4-Amino-4-Deoxy-L- Arabinose Biosynthesis.
J.Biol.Chem., 280, 2005
|
|
7AB9
 
 | | Crystal structure of human phosphodiesterase 4D2 catalytic domain with inhibitor NPD-656 | | Descriptor: | 1,2-ETHANEDIOL, 1-cycloheptyl-3-{4-methoxy-3-[2-(4-methoxyphenyl)ethoxy]phenyl}-4,4-dimethyl-4,5-dihydro-1H-pyrazol-5-one, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Singh, A.K, Blaazer, A.R, Zara, L, de Esch, I.J.P, Leurs, R, Brown, D.G. | | Deposit date: | 2020-09-07 | | Release date: | 2021-10-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | hPDE4D2 structure with inhibitor NPD-656
To be published
|
|
2BZR
 
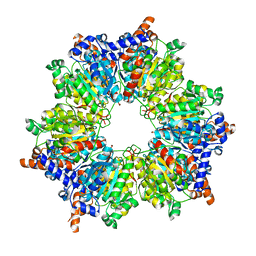 | |
2C4E
 
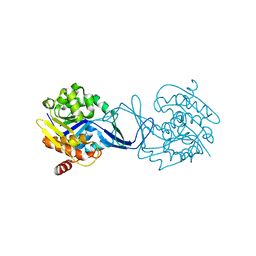 | | Crystal Structure of Methanocaldococcus jannaschii Nucleoside Kinase - An Archaeal Member of the Ribokinase Family | | Descriptor: | MAGNESIUM ION, SUGAR KINASE MJ0406 | | Authors: | Arnfors, L, Hansen, T, Meining, W, Schoenheit, P, Ladenstein, R. | | Deposit date: | 2005-10-18 | | Release date: | 2006-08-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of Methanocaldococcus Jannaschii Nucleoside Kinase: An Archaeal Member of the Ribokinase Family.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
7ABJ
 
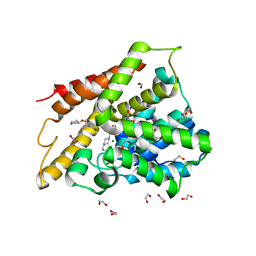 | | Crystal structure of human phosphodiesterase 4D2 catalytic domain with inhibitor NPD-1361 | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 5-(3-(cyclopentyloxy)-4-methoxyphenyl)-2-isopropyl-4,4-dimethyl-2,4-dihydro-3H-pyrazol-3-one, ... | | Authors: | Singh, A.K, Blaazer, A.R, Zara, L, de Esch, I.J.P, Leurs, R, Brown, D.G. | | Deposit date: | 2020-09-07 | | Release date: | 2021-10-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | hPDE4D2 structure with inhibitor NPD-1361
To be published
|
|
2C3F
 
 | | The structure of a group A streptococcal phage-encoded tail-fibre showing hyaluronan lyase activity. | | Descriptor: | HYALURONIDASE, PHAGE ASSOCIATED, SODIUM ION | | Authors: | Taylor, E.J, Smith, N.L, Linsay, A.-M, Charnock, S.J, Turkenburg, J.P, Dodson, E.J, Davies, G.J, Black, G.W. | | Deposit date: | 2005-10-06 | | Release date: | 2005-11-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structure of a Group a Streptococcal Phage-Encoded Virulence Factor Reveals Catalytically Active Triple-Stranded Beta-Helix
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
2C26
 
 | | Structural basis for the promiscuous specificity of the carbohydrate- binding modules from the beta-sandwich super family | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, ENDOGLUCANASE | | Authors: | Najmudin, S, Guerreiro, C.I.P.D, Carvalho, A.L, Bolam, D.N, Prates, J.A.M, Correia, M.A.S, Alves, V.D, Ferreira, L.M.A, Romao, M.J, Gilbert, H.J, Fontes, C.M.G.A. | | Deposit date: | 2005-09-26 | | Release date: | 2005-10-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Xyloglucan is Recognized by Carbohydrate-Binding Modules that Interact with Beta-Glucan Chains.
J.Biol.Chem., 281, 2006
|
|
2C6A
 
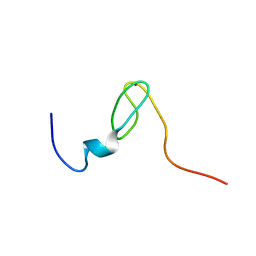 | | Solution structure of the C4 zinc-finger domain of HDM2 | | Descriptor: | UBIQUITIN-PROTEIN LIGASE E3 MDM2, ZINC ION | | Authors: | Yu, G.W, Allen, M.D, Andreeva, A, Fersht, A.R, Bycroft, M. | | Deposit date: | 2005-11-08 | | Release date: | 2006-01-04 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the C4 Zinc Finger Domain of Hdm2.
Protein Sci., 15, 2006
|
|
2BIL
 
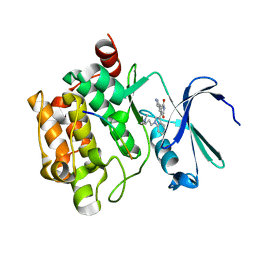 | | The human protein kinase Pim1 in complex with its consensus peptide Pimtide | | Descriptor: | 3-{1-[3-(DIMETHYLAMINO)PROPYL]-1H-INDOL-3-YL}-4-(1H-INDOL-3-YL)-1H-PYRROLE-2,5-DIONE, CONSENSUS PIM1 PEPTIDE PIMTIDE, PROTO-ONCOGENE SERINE/THREONINE-PROTEIN KINASE PIM-1 | | Authors: | Knapp, S, Debreczeni, J, Bullock, A, von Delft, F, Sundstrom, M, Arrowsmith, C, Edwards, A, Guo, K. | | Deposit date: | 2005-01-22 | | Release date: | 2005-02-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | The Human Protein Kinase Pim1 in Complex with its Consensus Peptide Pimtide
To be Published
|
|
6CCL
 
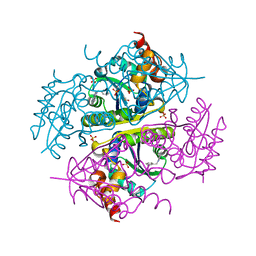 | | Crystal structure of E.coli Phosphopantetheine Adenylyltransferase (PPAT/CoaD) in complex with 1-benzyl-1H-imidazo[4,5-b]pyridine | | Descriptor: | 1-benzyl-1H-imidazo[4,5-b]pyridine, DIMETHYL SULFOXIDE, Phosphopantetheine adenylyltransferase, ... | | Authors: | Mamo, M, Appleton, B.A. | | Deposit date: | 2018-02-07 | | Release date: | 2018-03-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Fragment-Based Drug Discovery of Inhibitors of Phosphopantetheine Adenylyltransferase from Gram-Negative Bacteria.
J. Med. Chem., 61, 2018
|
|
2BG7
 
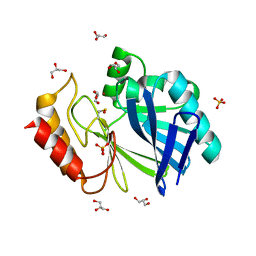 | | Bacillus cereus metallo-beta-lactamase (BcII) Arg (121) Cys mutant. Solved at pH4.5 using 20 Micromolar ZnSO4 in the buffer. 1mM DTT was used as a reducing agent. Cys221 is oxidized. | | Descriptor: | BETA-LACTAMASE II, GLYCEROL, SULFATE ION, ... | | Authors: | Davies, A.M, Rasia, R.M, Vila, A.J, Sutton, B.J, Fabiane, S.M. | | Deposit date: | 2004-12-17 | | Release date: | 2005-03-31 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Effect of Ph on the Active Site of an Arg121Cys Mutant of the Metallo-Beta-Lactamase from Bacillus Cereus: Implications for the Enzyme Mechanism
Biochemistry, 44, 2005
|
|
6CVP
 
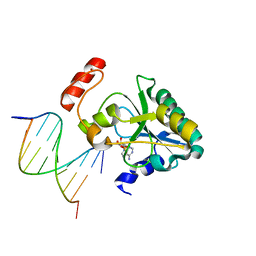 | | Human Aprataxin (Aptx) R199H bound to RNA-DNA, AMP and Zn product complex | | Descriptor: | ADENOSINE MONOPHOSPHATE, Aprataxin, DNA (5'-D(*GP*AP*AP*TP*CP*AP*TP*AP*AP*C)-3'), ... | | Authors: | Schellenberg, M.J, Williams, R.S, Tumbale, P.S. | | Deposit date: | 2018-03-28 | | Release date: | 2018-07-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.999 Å) | | Cite: | Mechanism of APTX nicked DNA sensing and pleiotropic inactivation in neurodegenerative disease.
EMBO J., 37, 2018
|
|
2BJC
 
 | | NMR structure of a protein-DNA complex of an altered specificity mutant of the lac repressor headpiece that mimics the gal repressor | | Descriptor: | 5'-D(*GP*AP*AP*TP*TP*GP*TP*AP*AP*GP *CP*GP*CP*TP*TP*AP*CP*AP*AP*TP*TP*C)-3', LACTOSE OPERON REPRESSOR | | Authors: | Salinas, R.K, Folkers, G.E, Bonvin, A.M.J.J, Das, D, Boelens, R, Kaptein, R. | | Deposit date: | 2005-02-01 | | Release date: | 2005-10-18 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | Altered Specificity in DNA Binding by the Lac Repressor: A Mutant Lac Headpiece that Mimics the Gal Repressor
Chembiochem, 6, 2005
|
|
2BN6
 
 | | P-Element Somatic Inhibitor Protein | | Descriptor: | PSI | | Authors: | Ignjatovic, T, Yang, J.C, Butler, P.J.G, Neuhaus, D, Nagai, K. | | Deposit date: | 2005-03-21 | | Release date: | 2005-07-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Basis of the Interaction between P-Element Somatic Inhibitor and U1-70K Essential for the Alternative Splicing of P-Element Transposase.
J.Mol.Biol., 351, 2005
|
|
