8DJ8
 
 | | Crystal Structure of Calgreen 1 protein | | Descriptor: | Calgreen One | | Authors: | Hung, L.-W, Nguyen, H.B, Waldo, G. | | Deposit date: | 2022-06-30 | | Release date: | 2023-07-05 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and Function of Engineered Variants of GFP-type Green Fluorescent protein from Corynactis californica (tentative)
To Be Published
|
|
8DJ2
 
 | |
8DJ1
 
 | | Crystal structure of NavAb V126T as a basis for the human Nav1.7 Inherited Erythromelalgia S241T mutation | | Descriptor: | 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, Ion transport protein | | Authors: | Wisedchaisri, G, Gamal El-Din, T.M, Zheng, N, Catterall, W.A. | | Deposit date: | 2022-06-29 | | Release date: | 2023-04-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural basis for severe pain caused by mutations in the S4-S5 linkers of voltage-gated sodium channel Na V 1.7.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8DJ0
 
 | | Crystal structure of NavAb L123T as a basis for the human Nav1.7 Inherited Erythromelalgia I848T mutation | | Descriptor: | 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, Ion transport protein | | Authors: | Wisedchaisri, G, Gamal El-Din, T.M, Zheng, N, Catterall, W.A. | | Deposit date: | 2022-06-29 | | Release date: | 2023-04-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for severe pain caused by mutations in the S4-S5 linkers of voltage-gated sodium channel Na V 1.7.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8DIY
 
 | | Crystal structure of NavAb L101S as a basis for the human Nav1.7 Inherited Erythromelalgia F216S mutation | | Descriptor: | 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, Ion transport protein | | Authors: | Wisedchaisri, G, Gamal El-Din, T.M, Zheng, N, Catterall, W.A. | | Deposit date: | 2022-06-29 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural basis for severe pain caused by mutations in the voltage sensors of sodium channel NaV1.7.
J.Gen.Physiol., 155, 2023
|
|
8DIW
 
 | | Crystal structure of NavAb E96P as a basis for the human Nav1.7 Inherited Erythromelalgia S211P mutation | | Descriptor: | 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, Ion transport protein | | Authors: | Wisedchaisri, G, Gamal El-Din, T.M, Zheng, N, Catterall, W.A. | | Deposit date: | 2022-06-29 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Structural basis for severe pain caused by mutations in the voltage sensors of sodium channel NaV1.7.
J.Gen.Physiol., 155, 2023
|
|
8DIV
 
 | | Crystal structure of NavAb I22V as a basis for the human Nav1.7 Inherited Erythromelalgia I136V mutation | | Descriptor: | 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, Ion transport protein, ... | | Authors: | Wisedchaisri, G, Gamal El-Din, T.M, Powell, N.M, Zheng, N, Catterall, W.A. | | Deposit date: | 2022-06-29 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Structural basis for severe pain caused by mutations in the voltage sensors of sodium channel NaV1.7.
J.Gen.Physiol., 155, 2023
|
|
8DIQ
 
 | | Tubulin-RB3_SLD-TTL in complex with SB226 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 4-[2-(ethylamino)-6,7-dihydro-5H-cyclopenta[d]pyrimidin-4-yl]-7-methoxy-3,4-dihydroquinoxalin-2(1H)-one, CALCIUM ION, ... | | Authors: | White, S.W, Yun, M. | | Deposit date: | 2022-06-29 | | Release date: | 2023-01-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.395 Å) | | Cite: | SB226, an inhibitor of tubulin polymerization, inhibits paclitaxel-resistant melanoma growth and spontaneous metastasis.
Cancer Lett., 555, 2022
|
|
8DII
 
 | |
8DIH
 
 | | Virtual screening for novel SARS-CoV-2 main protease non-covalent and covalent inhibitors | | Descriptor: | (1P,1'R)-1-(isoquinolin-4-yl)-2',3'-dihydrospiro[imidazolidine-4,1'-indene]-2,5-dione, 3C-like proteinase nsp5 | | Authors: | Singh, I, Shoichet, B.K. | | Deposit date: | 2022-06-29 | | Release date: | 2023-06-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Large library docking for novel SARS-CoV-2 main protease non-covalent and covalent inhibitors.
Protein Sci., 32, 2023
|
|
8DIG
 
 | |
8DIF
 
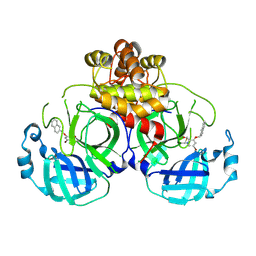 | |
8DIE
 
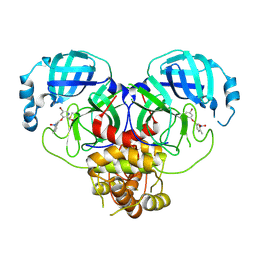 | |
8DID
 
 | |
8DIC
 
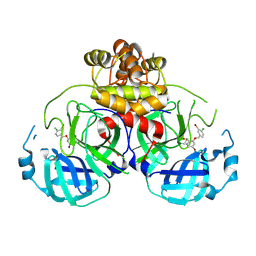 | |
8DIB
 
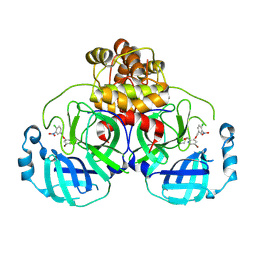 | |
8DI5
 
 | | Cryo-EM structure of SARS-CoV-2 Beta (B.1.351) spike protein in complex with VH domain F6 (focused refinement of RBD and VH F6) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, VH F6 | | Authors: | Zhu, X, Saville, J.W, Mannar, D, Berezuk, A.M, Subramaniam, S. | | Deposit date: | 2022-06-28 | | Release date: | 2022-08-24 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Potent and broad neutralization of SARS-CoV-2 variants of concern (VOCs) including omicron sub-lineages BA.1 and BA.2 by biparatopic human VH domains.
Iscience, 25, 2022
|
|
8DI4
 
 | | Discovery of MK-8189, a highly potent and selective PDE10A inhibitor for the treatment of schizophrenia | | Descriptor: | 2-methyl-6-{[(1S,2S)-2-(5-methylpyridin-2-yl)cyclopropyl]methoxy}-N-[(5-methyl-1,3,4-thiadiazol-2-yl)methyl]pyrimidin-4-amine, MAGNESIUM ION, ZINC ION, ... | | Authors: | Hayes, R.P, Yan, Y. | | Deposit date: | 2022-06-28 | | Release date: | 2023-02-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.016 Å) | | Cite: | Discovery of MK-8189, a Highly Potent and Selective PDE10A Inhibitor for the Treatment of Schizophrenia.
J.Med.Chem., 66, 2023
|
|
8DHT
 
 | | Crystal structure of a typeIII Rubisco | | Descriptor: | 3-PHOSPHOGLYCERIC ACID, ACETATE ION, GLYCEROL, ... | | Authors: | Qingqiu, H. | | Deposit date: | 2022-06-28 | | Release date: | 2022-11-30 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.699 Å) | | Cite: | Crystal structure of a type III Rubisco in complex with its product 3-phosphoglycerate.
Proteins, 91, 2023
|
|
8DHR
 
 | | An ester mutant of SfGFP | | Descriptor: | Green fluorescent protein | | Authors: | Reddi, R, Valiyaveetil, F.I. | | Deposit date: | 2022-06-28 | | Release date: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | A facile approach for incorporating tyrosine esters to probe ion-binding sites and backbone hydrogen bonds.
J.Biol.Chem., 300, 2023
|
|
8DHH
 
 | | DHODH IN COMPLEX WITH LIGAND 29 | | Descriptor: | (6M)-N-(3-chloro-2-methoxy-5-methylpyridin-4-yl)-6-[4-ethyl-3-(hydroxymethyl)-5-oxo-4,5-dihydro-1H-1,2,4-triazol-1-yl]-5-fluoro-2-{[(2S)-1,1,1-trifluoropropan-2-yl]oxy}pyridine-3-carboxamide, ACETATE ION, Dihydroorotate dehydrogenase (quinone), ... | | Authors: | Shaffer, P.L. | | Deposit date: | 2022-06-27 | | Release date: | 2022-08-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | N -Heterocyclic 3-Pyridyl Carboxamide Inhibitors of DHODH for the Treatment of Acute Myelogenous Leukemia.
J.Med.Chem., 65, 2022
|
|
8DHG
 
 | | DHODH IN COMPLEX WITH LIGAND 19 | | Descriptor: | (6M)-N-(2-chloro-4-methylpyridin-3-yl)-6-[4-ethyl-3-(hydroxymethyl)-5-oxo-4,5-dihydro-1H-1,2,4-triazol-1-yl]-5-fluoro-2-{[(2S)-1,1,1-trifluoropropan-2-yl]oxy}pyridine-3-carboxamide, ACETATE ION, Dihydroorotate dehydrogenase (quinone), ... | | Authors: | Shaffer, P.L. | | Deposit date: | 2022-06-27 | | Release date: | 2022-08-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | N -Heterocyclic 3-Pyridyl Carboxamide Inhibitors of DHODH for the Treatment of Acute Myelogenous Leukemia.
J.Med.Chem., 65, 2022
|
|
8DHF
 
 | | DHODH IN COMPLEX WITH LIGAND 11 | | Descriptor: | (6M)-N-(2-chloro-6-fluorophenyl)-6-[4-ethyl-3-(hydroxymethyl)-5-oxo-4,5-dihydro-1H-1,2,4-triazol-1-yl]-5-fluoro-2-{[(2S)-1,1,1-trifluoropropan-2-yl]oxy}pyridine-3-carboxamide, ACETATE ION, Dihydroorotate dehydrogenase (quinone), ... | | Authors: | Shaffer, P.L. | | Deposit date: | 2022-06-27 | | Release date: | 2022-08-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | N -Heterocyclic 3-Pyridyl Carboxamide Inhibitors of DHODH for the Treatment of Acute Myelogenous Leukemia.
J.Med.Chem., 65, 2022
|
|
8DHE
 
 | |
8DHA
 
 | |
