5SJD
 
 | | CRYSTAL STRUCTURE OF HUMAN PHOSPHODIESTERASE 10 IN COMPLEX WITH c1(ccnn1c2ccccc2)NC(=O)NCCn3c(cc(n3)C)C, micromolar IC50=>8.112831 | | Descriptor: | MAGNESIUM ION, N-[2-(3,5-dimethyl-1H-pyrazol-1-yl)ethyl]-N'-(1-phenyl-1H-pyrazol-5-yl)urea, ZINC ION, ... | | Authors: | Joseph, C, Benz, J, Flohr, A, Koerner, M, Rudolph, M.G. | | Deposit date: | 2022-02-01 | | Release date: | 2022-10-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Crystal Structure of a human phosphodiesterase 10 complex
To be published
|
|
4NUS
 
 | | Rsk2 N-terminal kinase in complex with LJH685 | | Descriptor: | 2,6-difluoro-4-{4-[4-(4-methylpiperazin-1-yl)phenyl]pyridin-3-yl}phenol, Ribosomal protein S6 kinase alpha-3 | | Authors: | Appleton, B.A. | | Deposit date: | 2013-12-04 | | Release date: | 2014-03-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Novel potent and selective inhibitors of p90 ribosomal S6 kinase reveal the heterogeneity of RSK function in MAPK-driven cancers.
Mol Cancer Res, 12, 2014
|
|
1TZG
 
 | | Crystal structure of HIV-1 neutralizing human Fab 4E10 in complex with a 13-residue peptide containing the 4E10 epitope on gp41 | | Descriptor: | Envelope polyprotein GP160, Fab 4E10, GLYCEROL | | Authors: | Cardoso, R.M.F, Zwick, M.B, Stanfield, R.L, Kunert, R, Binley, J.M, Katinger, H, Burton, D.R, Wilson, I.A. | | Deposit date: | 2004-07-09 | | Release date: | 2005-03-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Broadly Neutralizing Anti-HIV Antibody 4E10 Recognizes a Helical Conformation of a Highly Conserved Fusion-Associated Motif in gp41
Immunity, 22, 2005
|
|
4IE2
 
 | | Crystal structure of human Arginase-2 complexed with inhibitor 1h | | Descriptor: | Arginase-2, mitochondrial, BENZAMIDINE, ... | | Authors: | Cousido-Siah, A, Mitschler, A, Ruiz, F.X, Beckett, P, Van Zandt, M.C, Ji, M.K, Whitehouse, D, Ryder, T, Jagdmann, E, Andreoli, M, Mazur, A, Padmanilayam, M, Schroeter, H, Golebiowski, A, Podjarny, A. | | Deposit date: | 2012-12-13 | | Release date: | 2013-03-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2082 Å) | | Cite: | 2-Substituted-2-amino-6-boronohexanoic acids as arginase inhibitors.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
5SF1
 
 | | CRYSTAL STRUCTURE OF HUMAN PHOSPHODIESTERASE 10 IN COMPLEX WITH c1cc(nn2c1nc(c2C)C(F)F)C#Cc3nc(cn3C)c4ccccc4, micromolar IC50=0.001029 | | Descriptor: | (4S)-2-(difluoromethyl)-3-methyl-6-[(1-methyl-4-phenyl-1H-imidazol-2-yl)ethynyl]imidazo[1,2-b]pyridazine, MAGNESIUM ION, ZINC ION, ... | | Authors: | Joseph, C, Groebke-Zbinden, K, Benz, J, Schlatter, D, Rudolph, M.G. | | Deposit date: | 2022-01-21 | | Release date: | 2022-10-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | A high quality, industrial data set for binding affinity prediction: performance comparison in different early drug discovery scenarios.
J.Comput.Aided Mol.Des., 36, 2022
|
|
6PWD
 
 | | Ewingella americana HopBF1 kinase | | Descriptor: | 1,2-ETHANEDIOL, SULFATE ION, Type III effector HopBF1 | | Authors: | Tomchick, D.R, Tagliabracci, V.S, Park, B.C. | | Deposit date: | 2019-07-22 | | Release date: | 2019-09-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | A Bacterial Effector Mimics a Host HSP90 Client to Undermine Immunity.
Cell, 179, 2019
|
|
1PSH
 
 | |
6PWG
 
 | | Ewingella americana HopBF1 kinase bound to AMP-PNP | | Descriptor: | 1,2-ETHANEDIOL, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Type III effector HopBF1 | | Authors: | Tomchick, D.R, Tagliabracci, V.S, Park, B.C. | | Deposit date: | 2019-07-23 | | Release date: | 2019-09-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | A Bacterial Effector Mimics a Host HSP90 Client to Undermine Immunity.
Cell, 179, 2019
|
|
5BSU
 
 | | Crystal structure of 4-coumarate:CoA ligase complexed with caffeoyl adenylate | | Descriptor: | 4-coumarate--CoA ligase 2, 5'-O-[(R)-{[(2E)-3-(3,4-dioxocyclohexa-1,5-dien-1-yl)prop-2-enoyl]oxy}(hydroxy)phosphoryl]adenosine, GLYCEROL, ... | | Authors: | Li, Z, Nair, S.K. | | Deposit date: | 2015-06-02 | | Release date: | 2016-05-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Basis for Specificity and Flexibility in a Plant 4-Coumarate:CoA Ligase.
Structure, 23, 2015
|
|
5SGF
 
 | | CRYSTAL STRUCTURE OF HUMAN PHOSPHODIESTERASE 10 IN COMPLEX WITH c1(c(C(OCC)=O)cnn1C)C(NCCC)=O, micromolar IC50=6.982255 | | Descriptor: | GLYCEROL, MAGNESIUM ION, ZINC ION, ... | | Authors: | Joseph, C, Benz, J, Flohr, A, Peters, J, Rudolph, M.G. | | Deposit date: | 2022-02-01 | | Release date: | 2022-10-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of a human phosphodiesterase 10 complex
To be published
|
|
3H67
 
 | | Catalytic domain of human Serine/Threonine Phosphatase 5 (PP5c)with two Zn2+ atoms complexed with cantharidic acid | | Descriptor: | (1R,2S,3R,4S)-2,3-dimethyl-7-oxabicyclo[2.2.1]heptane-2,3-dicarboxylic acid, Serine/threonine-protein phosphatase 5, ZINC ION | | Authors: | Bertini, I, Calderone, V, Fragai, M, Luchinat, C, Talluri, E. | | Deposit date: | 2009-04-23 | | Release date: | 2009-09-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural basis of serine/threonine phosphatase inhibition by the archetypal small molecules cantharidin and norcantharidin
J.Med.Chem., 52, 2009
|
|
5SHB
 
 | | CRYSTAL STRUCTURE OF HUMAN PHOSPHODIESTERASE 10 IN COMPLEX WITH C2(=NN(c1ccncc1)C=CC2=O)c3ccnn3c4cccc(c4)Br, micromolar IC50=0.017329 | | Descriptor: | 3-[1-(3-bromophenyl)-1H-pyrazol-5-yl]-1-(pyridin-4-yl)pyridazin-4(1H)-one, MAGNESIUM ION, ZINC ION, ... | | Authors: | Joseph, C, Benz, J, Flohr, A, Koerner, M, Rudolph, M.G. | | Deposit date: | 2022-02-01 | | Release date: | 2022-10-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure of a human phosphodiesterase 10 complex
To be published
|
|
4IHP
 
 | | Crystal structure of TgCDPK1 with inhibitor bound | | Descriptor: | 1-tert-butyl-3-(3-chlorophenoxy)-1H-pyrazolo[3,4-d]pyrimidin-4-amine, Calmodulin-domain protein kinase 1, UNKNOWN ATOM OR ION | | Authors: | El Bakkouri, M, Tempel, W, Crandall, I.E, Massad, T, Loppnau, P, Graslund, S, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Kain, C.K, Shokat, K.M, Sibley, L.D, Hui, R, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-12-19 | | Release date: | 2014-01-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Crystal structure of TgCDPK1 with inhibitor bound
TO BE PUBLISHED
|
|
2P69
 
 | | Crystal Structure of Human Pyridoxal Phosphate Phosphatase with PLP | | Descriptor: | CALCIUM ION, PYRIDOXAL-5'-PHOSPHATE, Pyridoxal phosphate phosphatase | | Authors: | Ramagopal, U.A, Freeman, J, Izuka, M, Toro, R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-03-16 | | Release date: | 2007-04-03 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural genomics of protein phosphatases.
J.Struct.Funct.Genom., 8, 2007
|
|
3DJH
 
 | |
6JIR
 
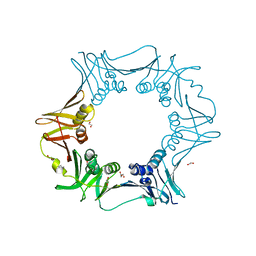 | | Crystal structure of C. crescentus beta sliding clamp with PEG bound to putative beta-motif tethering region | | Descriptor: | 1,2-ETHANEDIOL, Beta sliding clamp, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Jiang, X, Teng, M, Li, X. | | Deposit date: | 2019-02-23 | | Release date: | 2019-11-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Caulobacter crescentus beta sliding clamp employs a noncanonical regulatory model of DNA replication.
Febs J., 287, 2020
|
|
5SGB
 
 | | CRYSTAL STRUCTURE OF HUMAN PHOSPHODIESTERASE 10 IN COMPLEX WITH n3ncc(Oc1ccc(c2c1cccc2)Cl)s3, micromolar IC50=>10 | | Descriptor: | 5-[(4-chloronaphthalen-1-yl)oxy]-1,2,3-thiadiazole, MAGNESIUM ION, ZINC ION, ... | | Authors: | Joseph, C, Benz, J, Flohr, A, Brunner, M, Rudolph, M.G. | | Deposit date: | 2022-02-01 | | Release date: | 2022-10-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Crystal Structure of a human phosphodiesterase 10 complex
To be published
|
|
4YA1
 
 | | Yeast 20S proteasome beta2-H116N mutant | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Probable proteasome subunit alpha type-7, ... | | Authors: | Huber, E.M, Groll, M. | | Deposit date: | 2015-02-17 | | Release date: | 2015-06-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Systematic Analyses of Substrate Preferences of 20S Proteasomes Using Peptidic Epoxyketone Inhibitors.
J.Am.Chem.Soc., 137, 2015
|
|
3P1T
 
 | |
5SJA
 
 | | CRYSTAL STRUCTURE OF HUMAN PHOSPHODIESTERASE 10 IN COMPLEX WITH c13c(cc(s1)C(Nc2cc(nn2C(C)(C)C)C)=O)c(nn3c4ccccc4)COc5ncccc5, micromolar IC50=0.075134 | | Descriptor: | MAGNESIUM ION, N-(1-tert-butyl-3-methyl-1H-pyrazol-5-yl)-1-phenyl-3-{[(pyridin-2-yl)oxy]methyl}-1H-thieno[2,3-c]pyrazole-5-carboxamide, ZINC ION, ... | | Authors: | Joseph, C, Benz, J, Flohr, A, Koerner, M, Rudolph, M.G. | | Deposit date: | 2022-02-01 | | Release date: | 2022-10-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Crystal Structure of a human phosphodiesterase 10 complex
To be published
|
|
3H63
 
 | | Catalytic domain of human Serine/Threonine Phosphatase 5 (PP5c) with two Mn2+ atoms originally soaked with cantharidin (which is present in the structure in the hydrolyzed form) | | Descriptor: | (1R,2S,3R,4S)-2,3-dimethyl-7-oxabicyclo[2.2.1]heptane-2,3-dicarboxylic acid, MANGANESE (II) ION, Serine/threonine-protein phosphatase 5 | | Authors: | Bertini, I, Calderone, V, Fragai, M, Luchinat, C, Talluri, E. | | Deposit date: | 2009-04-23 | | Release date: | 2009-09-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural basis of serine/threonine phosphatase inhibition by the archetypal small molecules cantharidin and norcantharidin
J.Med.Chem., 52, 2009
|
|
7B8R
 
 | | Doxycycline bound structure of bacterial efflux pump. | | Descriptor: | (4S,4AR,5S,5AR,6R,12AS)-4-(DIMETHYLAMINO)-3,5,10,12,12A-PENTAHYDROXY-6-METHYL-1,11-DIOXO-1,4,4A,5,5A,6,11,12A-OCTAHYDROTETRACENE-2-CARBOXAMIDE, DARPin, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Wilhelm, J, Sjuts, H, Pos, K.M. | | Deposit date: | 2020-12-13 | | Release date: | 2021-10-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and functional analysis of the promiscuous AcrB and AdeB efflux pumps suggests different drug binding mechanisms.
Nat Commun, 12, 2021
|
|
5SHO
 
 | | CRYSTAL STRUCTURE OF HUMAN PHOSPHODIESTERASE 10 IN COMPLEX WITH C(=O)(NC1CC1)c5c(C(Nc3cc2nc(cn2cc3)c4ccccc4)=O)n(nc5)C, micromolar IC50=0.000960 | | Descriptor: | MAGNESIUM ION, N~4~-cyclopropyl-1-methyl-N~5~-[(4R)-2-phenylimidazo[1,2-a]pyridin-7-yl]-1H-pyrazole-4,5-dicarboxamide, ZINC ION, ... | | Authors: | Joseph, C, Benz, J, Flohr, A, Peters, J, Rudolph, M.G. | | Deposit date: | 2022-02-01 | | Release date: | 2022-10-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of a human phosphodiesterase 10 complex
To be published
|
|
5SHZ
 
 | | CRYSTAL STRUCTURE OF HUMAN PHOSPHODIESTERASE 10 IN COMPLEX WITH C2(=NN(c1ccccc1)C=CC2=O)c3ccnn3c4cccc(c4)C#C, micromolar IC50=0.004957 | | Descriptor: | 3-[1-(3-ethynylphenyl)-1H-pyrazol-5-yl]-1-phenylpyridazin-4(1H)-one, MAGNESIUM ION, ZINC ION, ... | | Authors: | Joseph, C, Benz, J, Flohr, A, Koerner, M, Rudolph, M.G. | | Deposit date: | 2022-02-01 | | Release date: | 2022-10-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal Structure of a human phosphodiesterase 10 complex
To be published
|
|
7XCR
 
 | | Cryo-EM structure of Dot1L and H2BK34ub-H3K79Nle nucleosome 1:1 complex | | Descriptor: | DNA (146-MER), Histone H2A, Histone H2B type 1-K, ... | | Authors: | Ai, H.S, Liu, A.J, Lou, Z.Y, Liu, L. | | Deposit date: | 2022-03-25 | | Release date: | 2022-04-20 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (2.57 Å) | | Cite: | H2B Lys34 Ubiquitination Induces Nucleosome Distortion to Stimulate Dot1L Activity.
Nat.Chem.Biol., 18, 2022
|
|
