7EGM
 
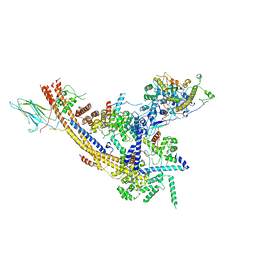 | | The SRM module of SWI/SNF-nucleosome complex | | Descriptor: | SWI/SNF chromatin-remodeling complex subunit SNF5, SWI/SNF chromatin-remodeling complex subunit SWI1, SWI/SNF complex subunit SWI3, ... | | Authors: | Chen, Z.C, Chen, K.J, He, Z.Y, Ye, Y.P. | | Deposit date: | 2021-03-24 | | Release date: | 2022-01-12 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure of the SWI/SNF complex bound to the nucleosome and insights into the functional modularity.
Cell Discov, 7, 2021
|
|
6O6Q
 
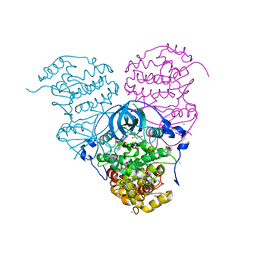 | | Crystal structure of Cka1p, a casein kinase 2 alpha ortholog from Candida albicans | | Descriptor: | ADENOSINE MONOPHOSPHATE, CHLORIDE ION, Casein kinase 2 catalytic subunit, ... | | Authors: | Stogios, P.J, Evdokimova, E, Di Leo, R, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-03-07 | | Release date: | 2019-04-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of Cka1p, a casein kinase 2 alpha ortholog from Candida albicans
To Be Published
|
|
6YW6
 
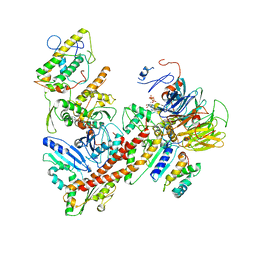 | | Cryo-EM structure of the ARP2/3 1B5CL isoform complex. | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ARPC1B, Actin-related protein 2, ... | | Authors: | von Loeffelholz, O, Moores, C, Purkiss, A. | | Deposit date: | 2020-04-29 | | Release date: | 2020-07-22 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Cryo-EM of human Arp2/3 complexes provides structural insights into actin nucleation modulation by ARPC5 isoforms.
Biol Open, 9, 2020
|
|
6YW7
 
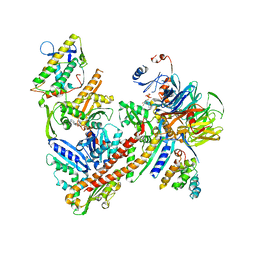 | | Cryo-EM structure of the ARP2/3 1A5C isoform complex. | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin-related protein 2, Actin-related protein 2/3 complex subunit 1A, ... | | Authors: | von Loeffelholz, O, Moores, C, Purkiss, A. | | Deposit date: | 2020-04-29 | | Release date: | 2020-07-22 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Cryo-EM of human Arp2/3 complexes provides structural insights into actin nucleation modulation by ARPC5 isoforms.
Biol Open, 9, 2020
|
|
9B8U
 
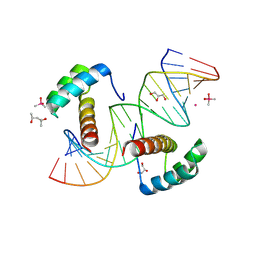 | |
3DT1
 
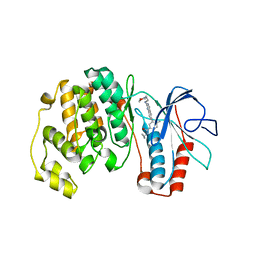 | | P38 Complexed with a quinazoline inhibitor | | Descriptor: | Mitogen-activated protein kinase 14, N-cyclopropyl-4-methyl-3-{2-[(2-morpholin-4-ylethyl)amino]quinazolin-6-yl}benzamide | | Authors: | Herberich, B, Syed, R, Li, V, Tasker, A.S. | | Deposit date: | 2008-07-14 | | Release date: | 2008-10-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery of highly selective and potent p38 inhibitors based on a phthalazine scaffold.
J.Med.Chem., 51, 2008
|
|
5L3C
 
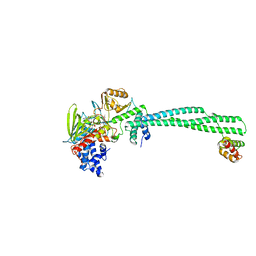 | | Human LSD1/CoREST: LSD1 E379K mutation | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Lysine-specific histone demethylase 1A, REST corepressor 1 | | Authors: | Pilotto, S, Speranzini, V, Marabelli, C, Mattevi, A. | | Deposit date: | 2016-04-06 | | Release date: | 2016-05-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.31 Å) | | Cite: | LSD1/KDM1A mutations associated to a newly described form of intellectual disability impair demethylase activity and binding to transcription factors.
Hum.Mol.Genet., 25, 2016
|
|
7B5O
 
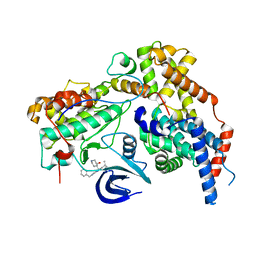 | | Cryo-EM structure of the human CAK bound to ICEC0942 at 2.5 Angstroms resolution | | Descriptor: | (3R,4R)-4-[[[7-[(phenylmethyl)amino]-3-propan-2-yl-pyrazolo[1,5-a]pyrimidin-5-yl]amino]methyl]piperidin-3-ol, CDK-activating kinase assembly factor MAT1, Cyclin-H, ... | | Authors: | Greber, B.J, Remis, J, Ali, S, Nogales, E. | | Deposit date: | 2020-12-05 | | Release date: | 2021-02-10 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | 2.5 angstrom -resolution structure of human CDK-activating kinase bound to the clinical inhibitor ICEC0942.
Biophys.J., 120, 2021
|
|
7B5S
 
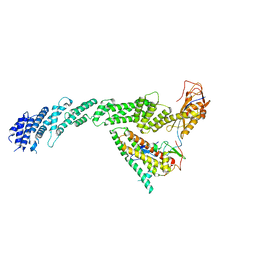 | | Ubiquitin ligation to F-box protein substrates by SCF-RBR E3-E3 super-assembly: CUL1-RBX1-ARIH1 Ariadne. Transition State 1 | | Descriptor: | Cullin-1, E3 ubiquitin-protein ligase ARIH1, E3 ubiquitin-protein ligase RBX1, ... | | Authors: | Horn-Ghetko, D, Prabu, J.R, Schulman, B.A. | | Deposit date: | 2020-12-07 | | Release date: | 2021-02-10 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Ubiquitin ligation to F-box protein targets by SCF-RBR E3-E3 super-assembly.
Nature, 590, 2021
|
|
7B5Q
 
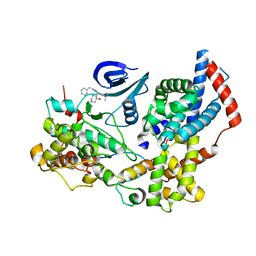 | | Cryo-EM structure of the human CAK bound to ICEC0942 (PHENIX-OPLS3e) | | Descriptor: | (3R,4R)-4-[[[7-[(phenylmethyl)amino]-3-propan-2-yl-pyrazolo[1,5-a]pyrimidin-5-yl]amino]methyl]piperidin-3-ol, CDK-activating kinase assembly factor MAT1, Cyclin-H, ... | | Authors: | Greber, B.J, Remis, J, Ali, S, Nogales, E. | | Deposit date: | 2020-12-05 | | Release date: | 2021-02-10 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | 2.5 angstrom -resolution structure of human CDK-activating kinase bound to the clinical inhibitor ICEC0942.
Biophys.J., 120, 2021
|
|
7WOT
 
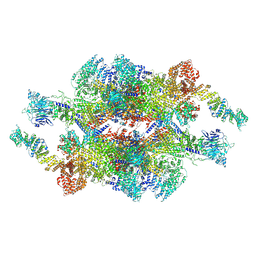 | | Cryo-EM structure of the inner ring monomer of the Saccharomyces cerevisiae nuclear pore complex | | Descriptor: | Nucleoporin NIC96, Nucleoporin NSP1, Nucleoporin NUP157, ... | | Authors: | Li, Z.Q, Chen, S.J.B, Zhao, L, Sui, S.F. | | Deposit date: | 2022-01-22 | | Release date: | 2022-04-13 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.73 Å) | | Cite: | Near-atomic structure of the inner ring of the Saccharomyces cerevisiae nuclear pore complex.
Cell Res., 32, 2022
|
|
7WOO
 
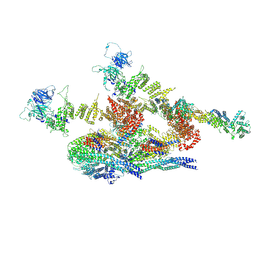 | | Cryo-EM structure of the inner ring protomer of the Saccharomyces cerevisiae nuclear pore complex | | Descriptor: | Nucleoporin NIC96, Nucleoporin NSP1, Nucleoporin NUP157, ... | | Authors: | Li, Z.Q, Chen, S.J.B, Zhao, L, Sui, S.F. | | Deposit date: | 2022-01-22 | | Release date: | 2022-04-13 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.71 Å) | | Cite: | Near-atomic structure of the inner ring of the Saccharomyces cerevisiae nuclear pore complex.
Cell Res., 32, 2022
|
|
7M1X
 
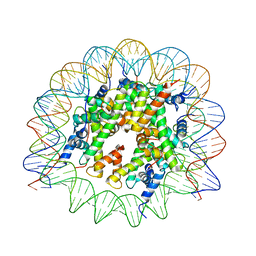 | |
5VHH
 
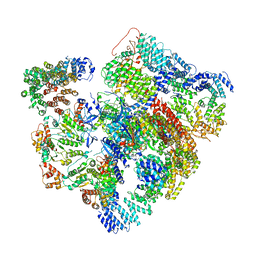 | | Conformational Landscape of the p28-Bound Human Proteasome Regulatory Particle | | Descriptor: | 26S proteasome complex subunit SEM1, 26S proteasome non-ATPase regulatory subunit 1, 26S proteasome non-ATPase regulatory subunit 10, ... | | Authors: | Lu, Y, Wu, J, Dong, Y, Chen, S, Sun, S, Ma, Y.B, Ouyang, Q, Finley, D, Kirschner, M.W, Mao, Y. | | Deposit date: | 2017-04-13 | | Release date: | 2017-08-23 | | Last modified: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (6.1 Å) | | Cite: | Conformational Landscape of the p28-Bound Human Proteasome Regulatory Particle.
Mol. Cell, 67, 2017
|
|
5VHI
 
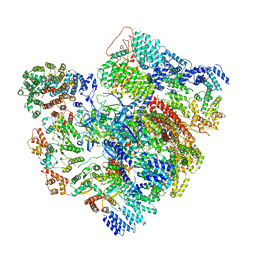 | | Conformational Landscape of the p28-Bound Human Proteasome Regulatory Particle | | Descriptor: | 26S proteasome complex subunit SEM1, 26S proteasome non-ATPase regulatory subunit 1, 26S proteasome non-ATPase regulatory subunit 10, ... | | Authors: | Lu, Y, Wu, J, Dong, Y, Chen, S, Sun, S, Ma, Y.B, Ouyang, Q, Finley, D, Kirschner, M.W, Mao, Y. | | Deposit date: | 2017-04-13 | | Release date: | 2017-08-23 | | Last modified: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (6.8 Å) | | Cite: | Conformational Landscape of the p28-Bound Human Proteasome Regulatory Particle.
Mol. Cell, 67, 2017
|
|
6WJD
 
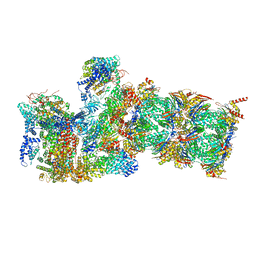 | |
8XVT
 
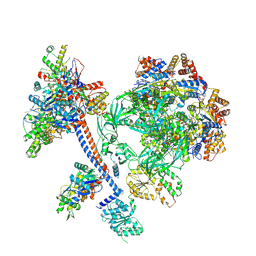 | | The core subcomplex of human NuA4/TIP60 complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, cytoplasmic 1, ... | | Authors: | Chen, K, Wang, L, Yu, Z, Yu, J, Ren, Y, Wang, Q, Xu, Y. | | Deposit date: | 2024-01-15 | | Release date: | 2024-07-24 | | Last modified: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure of the human TIP60 complex.
Nat Commun, 15, 2024
|
|
8XVG
 
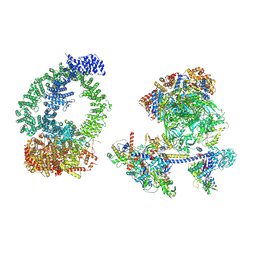 | | Structure of human NuA4/TIP60 complex | | Descriptor: | ACTB protein (Fragment), ADENOSINE-5'-DIPHOSPHATE, Actin-like protein 6A, ... | | Authors: | Chen, K, Wang, L, Yu, Z, Yu, J, Ren, Y, Wang, Q, Xu, Y. | | Deposit date: | 2024-01-15 | | Release date: | 2024-07-24 | | Last modified: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (9.4 Å) | | Cite: | Structure of the human TIP60 complex.
Nat Commun, 15, 2024
|
|
5LHG
 
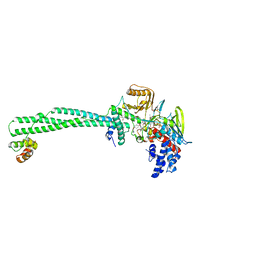 | | Structure of the KDM1A/CoREST complex with the inhibitor 4-methyl-N-[4-[[4-(1-methylpiperidin-4-yl)oxyphenoxy]methyl]phenyl]thieno[3,2-b]pyrrole-5-carboxamide | | Descriptor: | 4-methyl-N-[4-[[4-[(1-methyl-4-piperidyl)oxy]phenoxy]methyl]phenyl]thieno[3,2-b]pyrrole-5-carboxamide, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Cecatiello, V, Pasqualato, S. | | Deposit date: | 2016-07-11 | | Release date: | 2017-02-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.34 Å) | | Cite: | Thieno[3,2-b]pyrrole-5-carboxamides as New Reversible Inhibitors of Histone Lysine Demethylase KDM1A/LSD1. Part 2: Structure-Based Drug Design and Structure-Activity Relationship.
J. Med. Chem., 60, 2017
|
|
1W7B
 
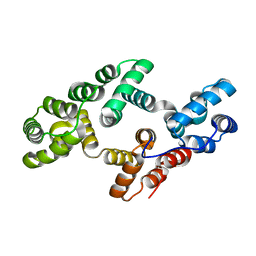 | |
3FLN
 
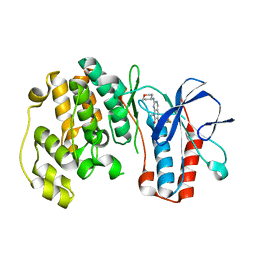 | | P38 kinase crystal structure in complex with R1487 | | Descriptor: | 6-(2,4-difluorophenoxy)-8-methyl-2-(tetrahydro-2H-pyran-4-ylamino)pyrido[2,3-d]pyrimidin-7(8H)-one, Mitogen-activated protein kinase 14 | | Authors: | Kuglstatter, A, Knapp, M. | | Deposit date: | 2008-12-19 | | Release date: | 2009-12-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Discovery of Pamapimod, R1503 and R1487 as Orally Bioavailable and Highly Selective Inhibitors of p38 Map Kinase
To be Published
|
|
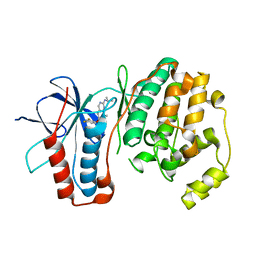
3FLY
 
 | |
5LHI
 
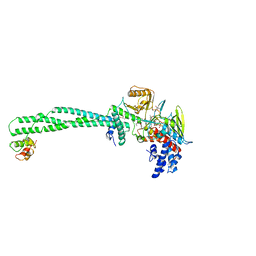 | | Structure of the KDM1A/CoREST complex with the inhibitor N-[3-(ethoxymethyl)-2-[[4-[[(3R)-pyrrolidin-3-yl]methoxy]phenoxy]methyl]phenyl]-4-methylthieno[3,2-b]pyrrole-5-carboxamide | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, LYSINE-SPECIFIC HISTONE DEMETHYLASE 1, ... | | Authors: | Cecatiello, V, Pasqualato, S. | | Deposit date: | 2016-07-12 | | Release date: | 2017-02-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Thieno[3,2-b]pyrrole-5-carboxamides as New Reversible Inhibitors of Histone Lysine Demethylase KDM1A/LSD1. Part 2: Structure-Based Drug Design and Structure-Activity Relationship.
J. Med. Chem., 60, 2017
|
|
3FLQ
 
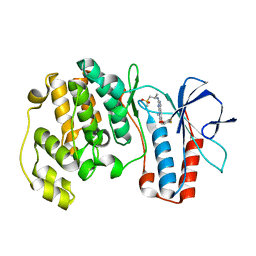 | |
5LHH
 
 | | Structure of the KDM1A/CoREST complex with the inhibitor 4-ethyl-N-[3-(methoxymethyl)-2-[[4-[[(3R)-pyrrolidin-3-yl]methoxy]phenoxy]methyl]phenyl]thieno[3,2-b]pyrrole-5-carboxamide | | Descriptor: | 4-ethyl-~{N}-[3-(methoxymethyl)-2-[[4-[[(3~{R})-pyrrolidin-3-yl]methoxy]phenoxy]methyl]phenyl]thieno[3,2-b]pyrrole-5-carboxamide, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Cecatiello, V, Pasqualato, S. | | Deposit date: | 2016-07-11 | | Release date: | 2017-02-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Thieno[3,2-b]pyrrole-5-carboxamides as New Reversible Inhibitors of Histone Lysine Demethylase KDM1A/LSD1. Part 2: Structure-Based Drug Design and Structure-Activity Relationship.
J. Med. Chem., 60, 2017
|
|
