7VE0
 
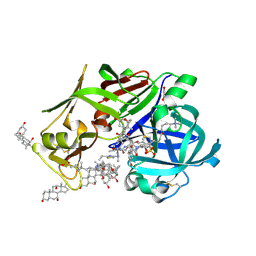 | | Crystal Structure of Ritonavir bound Plasmepsin II (PMII) from Plasmodium falciparum | | Descriptor: | 1,2-ETHANEDIOL, 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, Plasmepsin II, ... | | Authors: | Mishra, V, Rathore, I, Bhaumik, P. | | Deposit date: | 2021-09-07 | | Release date: | 2023-02-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Inhibition of Plasmodium falciparum plasmepsins by drugs targeting HIV-1 protease: A way forward for antimalarial drug discovery.
Curr Res Struct Biol, 7, 2024
|
|
9D3D
 
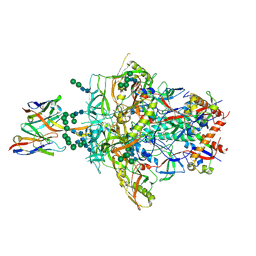 | | Cryo-EM structure of PGT145 R100aS Fab bound to HIV-1 BG505 DS-SOSIP.664 Env trimer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BG505 DS-SOSIP glycoprotein gp41, ... | | Authors: | Hodges, S, Morano, N.C, Shapiro, L, Kwong, P.D, Gorman, J. | | Deposit date: | 2024-08-09 | | Release date: | 2024-12-25 | | Last modified: | 2025-02-05 | | Method: | ELECTRON MICROSCOPY (3.41 Å) | | Cite: | Structural development of the HIV-1 apex-directed PGT145-PGDM1400 antibody lineage.
Cell Rep, 44, 2025
|
|
5I92
 
 | |
9D1W
 
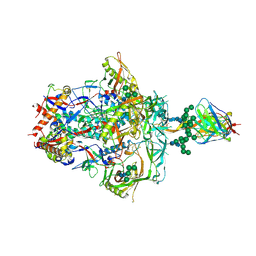 | | Cryo-EM structure of PGDM1400 Fab bound to HIV-1 BG505 DS-SOSIP.664 Env trimer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, HIV-1 BG505 DS-SOSIP glycoprotein gp41, ... | | Authors: | Kanai, T, Morano, N.C, Shapiro, L, Kwong, P.D, Gorman, J. | | Deposit date: | 2024-08-08 | | Release date: | 2024-12-25 | | Last modified: | 2025-02-05 | | Method: | ELECTRON MICROSCOPY (3.44 Å) | | Cite: | Structural development of the HIV-1 apex-directed PGT145-PGDM1400 antibody lineage.
Cell Rep, 44, 2025
|
|
7BG7
 
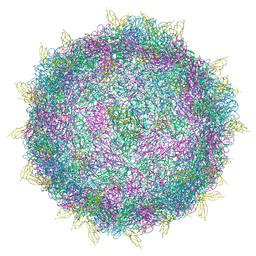 | | HRV14 in complex with its receptor ICAM-1 | | Descriptor: | Genome polyprotein, Intercellular adhesion molecule 1 | | Authors: | Hrebik, D, Fuzik, T, Plevka, P. | | Deposit date: | 2021-01-06 | | Release date: | 2021-05-19 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | ICAM-1 induced rearrangements of capsid and genome prime rhinovirus 14 for activation and uncoating.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7BEA
 
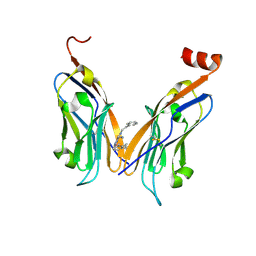 | | Structure of human Programmed cell death 1 ligand 1 (PD-L1) with inhibitor | | Descriptor: | 2-(aminomethyl)-6-[(2-methyl-3-phenyl-phenyl)methoxy]-~{N}-(2-phenylethyl)imidazo[1,2-a]pyridin-3-amine, Programmed cell death 1 ligand 1 | | Authors: | Magiera-Mularz, K, Butera, R, Wazynska, M, Holak, T, Domling, A. | | Deposit date: | 2020-12-22 | | Release date: | 2021-06-09 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Design, Synthesis, and Biological Evaluation of Imidazopyridines as PD-1/PD-L1 Antagonists.
Acs Med.Chem.Lett., 12, 2021
|
|
4H17
 
 | |
6C7D
 
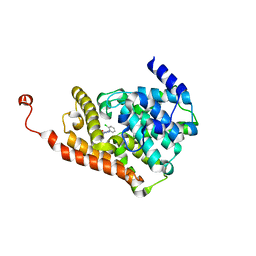 | | Crystal structure of human phosphodiesterase 2A with N-(1-adamantyl)-1-(2-chlorophenyl)-4-methyl-[1,2,4]triazolo[4,3-a]quinoxaline-8-carboxamide | | Descriptor: | 1-(2-chlorophenyl)-4-methyl-N-[(3s,5s,7s)-tricyclo[3.3.1.1~3,7~]decan-1-yl][1,2,4]triazolo[4,3-a]quinoxaline-8-carboxamide, MAGNESIUM ION, ZINC ION, ... | | Authors: | Xu, R, Aertgeerts, K. | | Deposit date: | 2018-01-22 | | Release date: | 2018-08-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Mathematical and Structural Characterization of Strong Nonadditive Structure-Activity Relationship Caused by Protein Conformational Changes.
J. Med. Chem., 61, 2018
|
|
4H5F
 
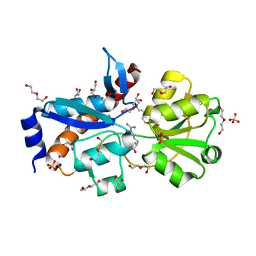 | | Crystal structure of an amino acid ABC transporter substrate-binding protein from Streptococcus pneumoniae Canada MDR_19A bound to L-arginine, form 1 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, ARGININE, ... | | Authors: | Stogios, P.J, Wawrzak, Z, Kudritska, M, Minasov, G, Yim, V, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-09-18 | | Release date: | 2012-10-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of an amino acid ABC transporter substrate-binding protein from Streptococcus pneumoniae Canada MDR_19A bound to L-arginine, form 1
TO BE PUBLISHED
|
|
2XNE
 
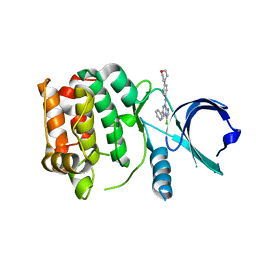 | | Structure of Aurora-A bound to an imidazopyrazine inhibitor | | Descriptor: | 3-chloro-N-(4-morpholin-4-ylphenyl)-6-pyridin-3-ylimidazo[1,2-a]pyrazin-8-amine, SERINE/THREONINE-PROTEIN KINASE 6 | | Authors: | Kosmopoulou, M, Bayliss, R. | | Deposit date: | 2010-08-02 | | Release date: | 2010-09-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure-based design of imidazo[1,2-a]pyrazine derivatives as selective inhibitors of Aurora-A kinase in cells.
Bioorg. Med. Chem. Lett., 20, 2010
|
|
7UX3
 
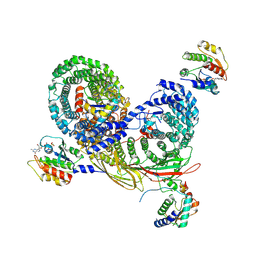 | | Asymmetric unit of AP-1, Arf1, Nef lattice on MHC-I lipopeptide incorporated narrow membrane tubes | | Descriptor: | ADP-ribosylation factor 1, AP-1 complex subunit beta-1, AP-1 complex subunit gamma-1, ... | | Authors: | Hooy, R.M, Hurley, J.H. | | Deposit date: | 2022-05-04 | | Release date: | 2023-05-10 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (9.6 Å) | | Cite: | Self-assembly and structure of a clathrin-independent AP-1:Arf1 tubular membrane coat.
Sci Adv, 8, 2022
|
|
9EHM
 
 | | Structure of HIV-1 BG505 SOSIP.664 Env trimer in complex with IOMAmin5 and 10-1074 Broadly Neutralizing Antibodies - Class II | | Descriptor: | 10-1074 Fab Heavy Chain, 10-1074 Fab Light Chain, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Dam, K.A, Yang, Z, Bjorkman, P.J. | | Deposit date: | 2024-11-23 | | Release date: | 2025-05-21 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Mapping essential somatic hypermutations in a CD4-binding site bNAb informs HIV-1 vaccine design.
Cell Rep, 44, 2025
|
|
4PUX
 
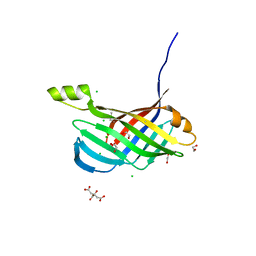 | |
9EHL
 
 | | Structure of HIV-1 BG505 SOSIP.664 Env trimer in complex with IOMAmin5 and 10-1074 Broadly Neutralizing Antibodies - Class I | | Descriptor: | 10-1074 Fab Heavy Chain, 10-1074 Fab Light Chain, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Dam, K.A, Yang, Z, Bjorkman, P.J. | | Deposit date: | 2024-11-23 | | Release date: | 2025-05-21 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Mapping essential somatic hypermutations in a CD4-binding site bNAb informs HIV-1 vaccine design.
Cell Rep, 44, 2025
|
|
5A86
 
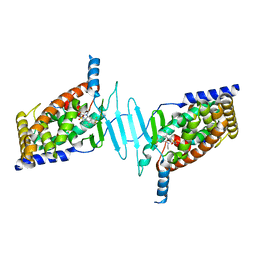 | | Structure of pregnane X receptor in complex with a Sphingosine 1- Phosphate Receptor 1 Antagonist | | Descriptor: | 4-chloro-N-[(1R)-1-[1-ethyl-6-(trifluoromethyl)benzimidazol-2-yl]ethyl]benzenesulfonamide, NUCLEAR RECEPTOR COACTIVATOR 1, NUCLEAR RECEPTOR SUBFAMILY 1 GROUP I MEMBER 2 | | Authors: | Xue, Y, Oster, L. | | Deposit date: | 2015-07-13 | | Release date: | 2015-10-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Identification and Optimization of Benzimidazole Sulfonamides as Orally Bioavailable Sphingosine 1-Phosphate Receptor 1 Antagonists with in Vivo Activity.
J.Med.Chem., 58, 2015
|
|
7KEP
 
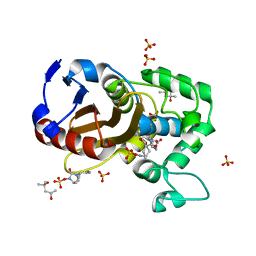 | | avibactam-CDD-1 2 minute complex | | Descriptor: | (2S,5R)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carboxamide, (2S,5R)-7-oxo-6-(sulfooxy)-1,6-diazabicyclo[3.2.1]octane-2-carboxamide, (4S)-2-METHYL-2,4-PENTANEDIOL, ... | | Authors: | Smith, C.A, Vakulenko, S.B, Stewart, N.K, Toth, M. | | Deposit date: | 2020-10-11 | | Release date: | 2021-01-20 | | Last modified: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Inhibition of the Clostridioides difficile Class D beta-Lactamase CDD-1 by Avibactam.
Acs Infect Dis., 7, 2021
|
|
4BZV
 
 | | The Solution Structure of the MLN 944-d(TACGCGTA)2 complex | | Descriptor: | 1-METHYL-9-[12-(9-METHYLPHENAZIN-10-IUM-1-YL)-12-OXO-2,11-DIAZA-5,8-DIAZONIADODEC-1-ANOYL]PHENAZIN-10-IUM, DNA | | Authors: | Serobian, A, Thomas, D.S, Ball, G.E, Denny, W.A, Wakelin, L.P.G. | | Deposit date: | 2013-07-30 | | Release date: | 2013-08-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The Solution Structure of Bis(Phenazine-1-Carboxamide)-DNA Complexes: Mln 944 Binding Corrected and Extended.
Biopolymers, 101, 2014
|
|
3KGW
 
 | |
3JQ1
 
 | |
7DCZ
 
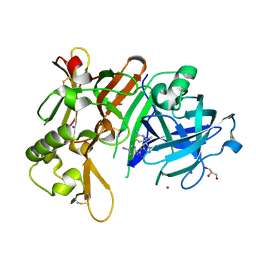 | | Crystal Structure of BACE1 in complex with N-{3-[(4S)-2-amino-4-methyl-4H-1,3-thiazin-4-yl]-4- fluorophenyl}-5-cyanopyridine-2-carboxamide | | Descriptor: | Beta-secretase 1, GLYCEROL, IODIDE ION, ... | | Authors: | Koriyama, Y, Hori, A, Ito, H, Yonezawa, S, Baba, Y, Tanimoto, N, Ueno, T, Yamamoto, S, Yamamoto, T, Asada, N, Morimoto, K, Einaru, S, Sakai, K, Kanazu, T, Matsuda, A, Yamaguchi, Y, Oguma, T, Timmers, M, Tritsmans, L, Kusakabe, K.I, Kato, A, Sakaguchi, G. | | Deposit date: | 2020-10-27 | | Release date: | 2021-03-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of Atabecestat (JNJ-54861911): A Thiazine-Based beta-Amyloid Precursor Protein Cleaving Enzyme 1 Inhibitor Advanced to the Phase 2b/3 EARLY Clinical Trial.
J.Med.Chem., 64, 2021
|
|
7UHF
 
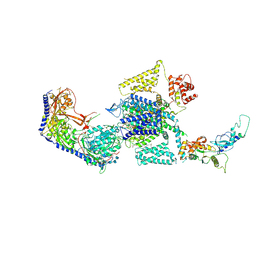 | | Human L-type voltage-gated calcium channel Cav1.3 in the presence of cinnarizine at 3.1 Angstrom resolution | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 1-(diphenylmethyl)-4-[(2E)-3-phenylprop-2-en-1-yl]piperazine, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gao, S, Yao, X, Yan, N. | | Deposit date: | 2022-03-26 | | Release date: | 2022-05-11 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis for pore blockade of human voltage-gated calcium channel Ca v 1.3 by motion sickness drug cinnarizine.
Cell Res., 32, 2022
|
|
7UGP
 
 | |
2AMF
 
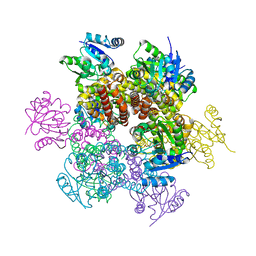 | | Crystal structure of 1-Pyrroline-5-Carboxylate Reductase from Human Pathogen Streptococcus Pyogenes | | Descriptor: | 1-Pyrroline-5-Carboxylate reductase, PROLINE, SODIUM ION | | Authors: | Nocek, B, Lezondra, L, Holzle, D, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-08-09 | | Release date: | 2005-09-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structures of Delta(1)-Pyrroline-5-carboxylate Reductase from Human Pathogens Neisseria meningitides and Streptococcus pyogenes
J.Mol.Biol., 354, 2005
|
|
6MEO
 
 | | Structural basis of coreceptor recognition by HIV-1 envelope spike | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Shaik, M.M, Chen, B. | | Deposit date: | 2018-09-06 | | Release date: | 2018-12-12 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural basis of coreceptor recognition by HIV-1 envelope spike.
Nature, 565, 2018
|
|
2XNG
 
 | | Structure of Aurora-A bound to a selective imidazopyrazine inhibitor | | Descriptor: | N-(3-{3-chloro-8-[(4-morpholin-4-ylphenyl)amino]imidazo[1,2-a]pyrazin-6-yl}benzyl)methanesulfonamide, SERINE/THREONINE-PROTEIN KINASE 6 | | Authors: | Kosmopoulou, M, Bayliss, R. | | Deposit date: | 2010-08-02 | | Release date: | 2010-09-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.605 Å) | | Cite: | Structure-based design of imidazo[1,2-a]pyrazine derivatives as selective inhibitors of Aurora-A kinase in cells.
Bioorg. Med. Chem. Lett., 20, 2010
|
|
