2RQZ
 
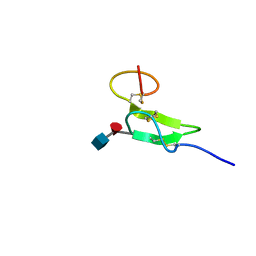 | | Structure of sugar modified epidermal growth factor-like repeat 12 of mouse Notch-1 receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-alpha-L-fucopyranose, Neurogenic locus notch homolog protein 1 | | Authors: | Shimizu, K, Fujitani, N, Hosoguchi, K, Nishimura, S. | | Deposit date: | 2010-02-26 | | Release date: | 2010-10-13 | | Last modified: | 2020-07-29 | | Method: | SOLUTION NMR | | Cite: | Chemical Synthesis, Folding, and Structural Insights into O-Fucosylated Epidermal Growth Factor-like Repeat 12 of Mouse Notch-1 Receptor
J.Am.Chem.Soc., 132, 2010
|
|
4FE8
 
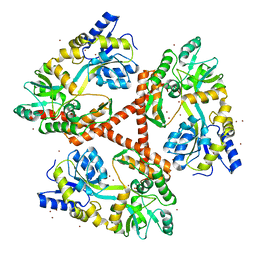 | | Crystal Structure of Htt36Q3H-EX1-X1-C1(Alpha) | | Descriptor: | Maltose-binding periplasmic protein,Huntingtin, ZINC ION | | Authors: | Kim, M. | | Deposit date: | 2012-05-29 | | Release date: | 2013-03-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Beta conformation of polyglutamine track revealed by a crystal structure of Huntingtin N-terminal region with insertion of three histidine residues.
Prion, 7, 2013
|
|
2RSC
 
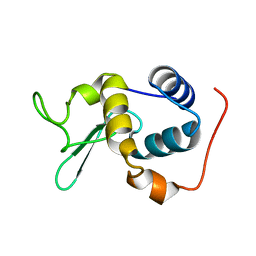 | |
3VFJ
 
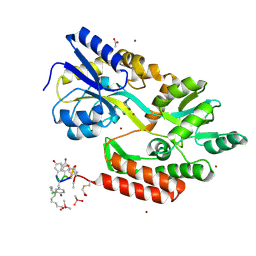 | | The structure of monodechloro-teicoplanin in complex with its ligand, using MBP as a ligand carrier | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-amino-2-deoxy-beta-D-glucopyranose, 8-METHYLNONANOIC ACID, ... | | Authors: | Economou, N.J, Weeks, S.D, Grasty, K.C, Loll, P.J. | | Deposit date: | 2012-01-09 | | Release date: | 2013-01-09 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure of the complex between teicoplanin and a bacterial cell-wall peptide: use of a carrier-protein approach.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
1PEB
 
 | | LIGAND-FREE HIGH-AFFINITY MALTOSE-BINDING PROTEIN | | Descriptor: | Maltose-binding periplasmic protein | | Authors: | Telmer, P.G, Shilton, B.H. | | Deposit date: | 2003-05-21 | | Release date: | 2003-08-12 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Insights into the Conformational Equilibria of Maltose-binding Protein by Analysis of High Affinity Mutants.
J.Biol.Chem., 278, 2003
|
|
8KGB
 
 | | SlNDPS1-AtcPT4 Chimera complexed with GSPP, Mg2+, and IPP | | Descriptor: | 3-METHYLBUT-3-ENYL TRIHYDROGEN DIPHOSPHATE, Dimethylallylcistransferase CPT1, chloroplastic,Dehydrodolichyl diphosphate synthase 2, ... | | Authors: | Suenaga-Hiromori, M, Ishii, T, Imaizumi, R, Takeshita, K, Yanai, T, Matsuura, H, Sakai, N, Yamaguchi, H, Yanbe, F, Waki, T, Tozawa, Y, Miyagi-Inoue, Y, Yamamoto, M, Kataoka, K, Nakayama, T, Yamashita, S, Takahashi, S. | | Deposit date: | 2023-08-18 | | Release date: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | A versatile system for enzymatic synthesis of natural and unnatural polyisoprenoids on rubber particles
To Be Published
|
|
8KGA
 
 | | SlNDPS1-AtcPT4 Chimera | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Dimethylallylcistransferase CPT1, ... | | Authors: | Suenaga-Hiromori, M, Ishii, T, Imaizumi, R, Takeshita, K, Yanai, T, Matsuura, H, Sakai, N, Yamaguchi, H, Yanbe, F, Waki, T, Tozawa, Y, Miyagi-Inoue, Y, Yamamoto, M, Kataoka, K, Nakayama, T, Yamashita, S, Takahashi, S. | | Deposit date: | 2023-08-18 | | Release date: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (2.182 Å) | | Cite: | Biosynthesising various rubber-like polymers by reconstituting prenyltransferases on Hevea rubber particles: molecular and structural bases of the versatile system
To Be Published
|
|
2BW0
 
 | | Crystal Structure of the hydrolase domain of Human 10-Formyltetrahydrofolate 2 dehydrogenase | | Descriptor: | 10-FORMYLTETRAHYDROFOLATE DEHYDROGENASE, SULFATE ION | | Authors: | Ogg, D.J, Stenmark, P, Arrowsmith, C, Edwards, A, Ehn, M, Graslund, S, Hammarstrom, M, Hallberg, M, Kotenyova, T, Nilsson-Ehle, P, Nordlund, P, Persson, C, Sagemark, J, Schuler, H, Sundstrom, M, Thorsell, A, Dobritzsch, D, Weigelt, J. | | Deposit date: | 2005-07-07 | | Release date: | 2005-07-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structures of the Hydrolase Domain of Human 10-Formyltetrahydrofolate Dehydrogenase and its Complex with a Substrate Analogue.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
3W15
 
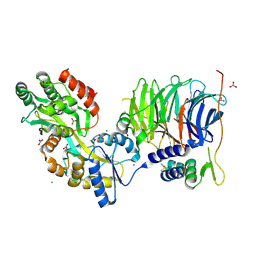 | | Structure of peroxisomal targeting signal 2 (PTS2) of Saccharomyces cerevisiae 3-ketoacyl-CoA thiolase in complex with Pex7p and Pex21p | | Descriptor: | 3-ketoacyl-CoA thiolase, peroxisomal, Maltose-binding periplasmic protein, ... | | Authors: | Pan, D, Nakatsu, T, Kato, H. | | Deposit date: | 2012-11-06 | | Release date: | 2013-07-03 | | Last modified: | 2017-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of peroxisomal targeting signal-2 bound to its receptor complex Pex7p-Pex21p
Nat.Struct.Mol.Biol., 20, 2013
|
|
2V93
 
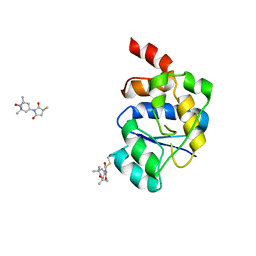 | | EQUILLIBRIUM MIXTURE OF OPEN AND PARTIALLY-CLOSED SPECIES IN THE APO STATE OF MALTODEXTRIN-BINDING PROTEIN BY PARAMAGNETIC RELAXATION ENHANCEMENT NMR | | Descriptor: | 1-(1-HYDROXY-2,2,6,6-TETRAMETHYLPIPERIDIN-4-YL)PYRROLIDINE-2,5-DIONE, MALTOSE-BINDING PERIPLASMIC PROTEIN | | Authors: | Clore, G.M, Tang, C. | | Deposit date: | 2007-08-21 | | Release date: | 2007-11-06 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Open-to-Closed Transition in Apo Maltose-Binding Protein Observed by Paramagnetic NMR.
Nature, 449, 2007
|
|
2VJ3
 
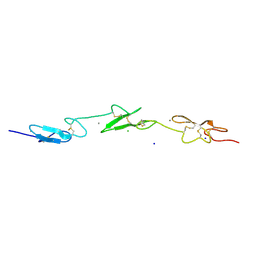 | | Human Notch-1 EGFs 11-13 | | Descriptor: | CALCIUM ION, CHLORIDE ION, NEUROGENIC LOCUS NOTCH HOMOLOG PROTEIN 1, ... | | Authors: | Johnson, S, Cordle, J, Tay, J.Z, Roversi, P, Lea, S.M. | | Deposit date: | 2007-12-06 | | Release date: | 2008-07-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A Conserved Face of the Jagged/Serrate Dsl Domain is Involved in Notch Trans-Activation and Cis-Inhibition.
Nat.Struct.Mol.Biol., 15, 2008
|
|
6FD5
 
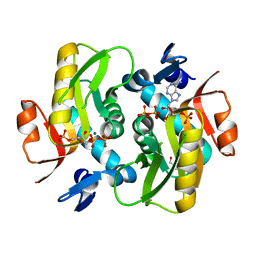 | | Crystal Structure of Human APRT-Tyr105Phe variant in complex with Adenine, PRPP and Mg2+, 14 days post crystallization (with AMP and PPi products partially generated) | | Descriptor: | 1-O-pyrophosphono-5-O-phosphono-alpha-D-ribofuranose, ADENOSINE MONOPHOSPHATE, Adenine phosphoribosyltransferase, ... | | Authors: | Nioche, P, Huyet, J, Ozeir, M. | | Deposit date: | 2017-12-21 | | Release date: | 2018-08-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural Insights into the Forward and Reverse Enzymatic Reactions in Human Adenine Phosphoribosyltransferase.
Cell Chem Biol, 25, 2018
|
|
2DTN
 
 | | Crystal structure of Helicobacter pylori undecaprenyl pyrophosphate synthase complexed with pyrophosphate | | Descriptor: | DIPHOSPHATE, undecaprenyl pyrophosphate synthase | | Authors: | Guo, R.T, Kuo, C.J, Chen, C.L, Ko, T.P, Liang, P.H, Wang, A.H.-J. | | Deposit date: | 2006-07-13 | | Release date: | 2007-06-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Biochemical characterization, crystal structure, and inhibitors of Helicobacter pylori undecaprenyl pyrophosphate synthase
To be Published
|
|
1N4C
 
 | | NMR Structure of the J-Domain and Clathrin Substrate Binding Domain of Bovine Auxilin | | Descriptor: | Auxilin | | Authors: | Gruschus, J.M, Han, C.J, Greener, T, Greene, L.E, Ferretti, J.A, Eisenberg, E. | | Deposit date: | 2002-10-30 | | Release date: | 2003-11-11 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of the functional fragment of auxilin required for catalytic uncoating of clathrin-coated vesicles.
Biochemistry, 43, 2004
|
|
6FD4
 
 | | Crystal Structure of Human APRT-Tyr105Phe variant in complex with Adenine, PRPP and Mg2+, 14 hours post crystallization | | Descriptor: | 1-O-pyrophosphono-5-O-phosphono-alpha-D-ribofuranose, Adenine phosphoribosyltransferase, MAGNESIUM ION | | Authors: | Nioche, P, Huyet, J, Ozeir, M. | | Deposit date: | 2017-12-21 | | Release date: | 2018-08-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Insights into the Forward and Reverse Enzymatic Reactions in Human Adenine Phosphoribosyltransferase.
Cell Chem Biol, 25, 2018
|
|
8OM3
 
 | | Small subunit of yeast mitochondrial ribosome in complex with IF3/Aim23. | | Descriptor: | 15S mitochondrial rRNA, 3-hydroxyisobutyryl-CoA hydrolase, mitochondrial, ... | | Authors: | Itoh, Y, Chicherin, I, Kamenski, P, Amunts, A. | | Deposit date: | 2023-03-31 | | Release date: | 2024-01-10 | | Last modified: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (2.87 Å) | | Cite: | METTL17 is an Fe-S cluster checkpoint for mitochondrial translation.
Mol.Cell, 84, 2024
|
|
6FCI
 
 | | Crystal Structure of Human APRT wild type in complex with Adenine, PRPP and Mg2+ | | Descriptor: | 1-O-pyrophosphono-5-O-phosphono-alpha-D-ribofuranose, ADENINE, Adenine phosphoribosyltransferase, ... | | Authors: | Nioche, P, Huyet, J, Ozeir, M. | | Deposit date: | 2017-12-20 | | Release date: | 2018-08-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structural Insights into the Forward and Reverse Enzymatic Reactions in Human Adenine Phosphoribosyltransferase.
Cell Chem Biol, 25, 2018
|
|
6FCH
 
 | |
7QSR
 
 | | CryoEM structure of the Ectodomain of Human PLA2R | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Secretory phospholipase A2 receptor, alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Briggs, D.C, Lockhart-Cairns, M.P, Baldock, C. | | Deposit date: | 2022-01-14 | | Release date: | 2022-07-20 | | Last modified: | 2022-08-03 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure of PLA2R reveals presentation of the dominant membranous nephropathy epitope and an immunogenic patch.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
2VRW
 
 | |
6FSW
 
 | | Structure of Archaeoglobus fulgidus SBDS protein at 1.9 Angstrom | | Descriptor: | DI(HYDROXYETHYL)ETHER, Ribosome maturation protein SDO1-like protein | | Authors: | Mazzorana, M, Foadi, J, Siliqi, D, Sanchez-Puig, N. | | Deposit date: | 2018-02-20 | | Release date: | 2018-03-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Conformational flexibility of proteins involved in ribosome biogenesis: investigations via Small Angle X-ray Scattering (SAXS)
Crystals, 8, 2018
|
|
2D2R
 
 | | Crystal structure of Helicobacter pylori Undecaprenyl Pyrophosphate Synthase | | Descriptor: | Undecaprenyl Pyrophosphate Synthase | | Authors: | Kuo, C.J, Guo, R.T, Chen, C.L, Ko, T.P, Cheng, Y.S, Cheng, Y.L, Liang, P.H, Wang, A.H.-J. | | Deposit date: | 2005-09-16 | | Release date: | 2006-09-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Structure-based inhibitors exhibit differential activities against Helicobacter pylori and Escherichia coli undecaprenyl pyrophosphate synthases.
J.Biomed.Biotechnol., 2008, 2008
|
|
1NMU
 
 | | MBP-L30 | | Descriptor: | 60S ribosomal protein L30, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, maltose-binding periplasmic protein | | Authors: | Chao, J.A, Prasad, G.S, White, S.A, Stout, C.D, Williamson, J.R. | | Deposit date: | 2003-01-10 | | Release date: | 2003-02-18 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Inherent Protein Structural Flexibility at the RNA-binding Interface of L30e
J.Mol.Biol., 326, 2003
|
|
8JXS
 
 | | Structure of nanobody-bound DRD1_PF-6142 complex | | Descriptor: | 4-[3-methyl-4-(6-methylimidazo[1,2-a]pyrazin-5-yl)phenoxy]furo[3,2-c]pyridine, D(1A) dopamine receptor, Fab 8D3 heavy chain, ... | | Authors: | Zhuang, Y, Xu, Y, Fan, L, Wang, S, Xu, H.E. | | Deposit date: | 2023-07-01 | | Release date: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of psychedelic LSD recognition at dopamine D 1 receptor.
Neuron, 2024
|
|
8JXR
 
 | | Structure of nanobody-bound DRD1_LSD complex | | Descriptor: | (8alpha)-N,N-diethyl-6-methyl-9,10-didehydroergoline-8-carboxamide, D(1A) dopamine receptor, Fab 8D3 heavy chain, ... | | Authors: | Zhuang, Y, Xu, Y, Fan, L, Wang, S, Xu, H.E. | | Deposit date: | 2023-07-01 | | Release date: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (3.57 Å) | | Cite: | Structural basis of psychedelic LSD recognition at dopamine D 1 receptor.
Neuron, 2024
|
|
