5UTO
 
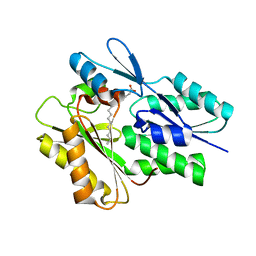 | | The crystal structure of the Staphylococcus aureus Fatty acid Kinase (Fak) B1 protein loaded with palmitic acid to 1.83 Angstrom resolution | | Descriptor: | EDD domain protein, DegV family, PALMITIC ACID | | Authors: | Cuypers, M.G, Ericson, M, Subramanian, C, Broussard, T.C, Miller, D.J, White, S.W, Rock, C.O. | | Deposit date: | 2017-02-15 | | Release date: | 2018-02-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | The crystal structure of the Staphylococcus aureus Fatty acid Kinase (Fak) B1 protein loaded with palmitic acid to 1.83 Angstroem resolution
J.Biol.Chem., 2018
|
|
7H4F
 
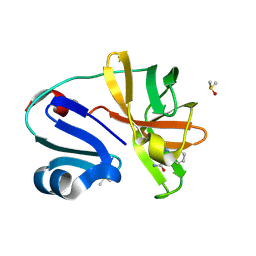 | | Group deposition for crystallographic fragment screening of Coxsackievirus A16 (G-10) 2A protease -- Crystal structure of Coxsackievirus A16 (G-10) 2A protease in complex with Z1618027593 (A71EV2A-x0812) | | Descriptor: | 1-methylindazole-3-carboxamide, DIMETHYL SULFOXIDE, Protease 2A, ... | | Authors: | Lithgo, R.M, Fairhead, M, Koekemoer, L, Balcomb, B.H, Capkin, E, Chandran, A.V, Golding, M, Godoy, A.S, Aschenbrenner, J.C, Marples, P.G, Ni, X, Thompson, W, Tomlinson, C.W.E, Wild, C, Winokan, M, Xavier, M.-A.E, Fearon, D, von Delft, F. | | Deposit date: | 2024-04-04 | | Release date: | 2024-04-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Crystallographic Fragment Screen of Coxsackievirus A16 2A Protease identifies new opportunities for the development of broad-spectrum anti-enterovirals.
Biorxiv, 2024
|
|
7JMG
 
 | | Functional Pathways of Biomolecules Retrieved from Single-particle Snapshots - Frame 22 - State 2 (S2) | | Descriptor: | CALCIUM ION, Peptidyl-prolyl cis-trans isomerase FKBP1B, ZINC ION, ... | | Authors: | Dashti, A, des Georges, A, Frank, J, Ourmazd, A. | | Deposit date: | 2020-07-31 | | Release date: | 2020-08-12 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Retrieving functional pathways of biomolecules from single-particle snapshots.
Nat Commun, 11, 2020
|
|
8X0I
 
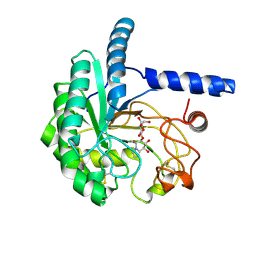 | | X-Ray crystal structure of glycoside hydrolase family 6 cellobiohydrolase from Phanerochaete chrysosporium PcCel6A C240S/C393S soaked in cellobioimidazole | | Descriptor: | (5R,6R,7R,8S)-7,8-dihydroxy-5-(hydroxymethyl)-5,6,7,8-tetrahydroimidazo[1,2-a]pyridin-6-yl beta-D-glucopyranoside, Glucanase | | Authors: | Yamaguchi, S, Sunagawa, N, Tachioka, M, Igarashi, K. | | Deposit date: | 2023-11-04 | | Release date: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Activity and X-ray crystal structure of candidate base catalyst mutants of glycoside hydrolase family 6 cellobiohydrolase
To Be Published
|
|
1L35
 
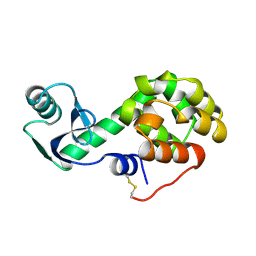 | |
7BHW
 
 | | Crystal structure of MAT2a bound to allosteric inhibitor (compound 29) | | Descriptor: | 7-chloranyl-4-(dimethylamino)-1-(3-methylphenyl)quinazolin-2-one, S-ADENOSYLMETHIONINE, S-adenosylmethionine synthase isoform type-2 | | Authors: | Schimpl, M, De Fusco, C, Borjesson, U, Cheung, T, Collie, I, Evans, L, Narasimhan, P, Stubbs, C, Vazquez-Chantada, M, Wagner, D.J, Grondine, M, Tentarelli, S, Underwood, E, Argyrou, A, Bagal, S, Chiarparin, E, Robb, G, Scott, J.S. | | Deposit date: | 2021-01-11 | | Release date: | 2021-04-21 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Fragment-Based Design of a Potent MAT2a Inhibitor and in Vivo Evaluation in an MTAP Null Xenograft Model.
J.Med.Chem., 64, 2021
|
|
6UKW
 
 | | STING C-terminal Domain Complexed with Non-cyclic Dinucleotide Compound 10 | | Descriptor: | 4-[6-(3-{[2-(3-carboxypropanoyl)-4-fluoro-6-methoxy-1-benzothiophen-5-yl]oxy}propoxy)-5-methoxy-1-benzothiophen-2-yl]-4-oxobutanoic acid, fusion protein of Ubiquitin-like protein SMT3 and Stimulator of interferon protein c-terminal domain | | Authors: | Lesburg, C.A. | | Deposit date: | 2019-10-06 | | Release date: | 2020-08-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | An orally available non-nucleotide STING agonist with antitumor activity.
Science, 369, 2020
|
|
9C1P
 
 | | Structure of Calcium-Sensing Receptor in complex with positive allosteric modulator '6218 | | Descriptor: | (5R)-N-[2-(1,2-benzothiazol-3-yl)ethyl]-1-methyl-2,3,4,5-tetrahydro-1H-1-benzazepin-5-amine, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Wu, C, Skiniotis, G. | | Deposit date: | 2024-05-29 | | Release date: | 2024-10-02 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Large library docking identifies positive allosteric modulators of the calcium-sensing receptor.
Science, 385, 2024
|
|
5VBM
 
 | | Crystal Structure of Small Molecule Disulfide 2C07 Bound to K-Ras Cys Light M72C GDP | | Descriptor: | 1-(4-methoxyphenyl)-N-(3-sulfanylpropyl)-5-(trifluoromethyl)-1H-pyrazole-4-carboxamide, GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Gentile, D.R, Jenkins, M.L, Moss, S.M, Burke, J.E, Shokat, K.M. | | Deposit date: | 2017-03-29 | | Release date: | 2017-10-25 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Ras Binder Induces a Modified Switch-II Pocket in GTP and GDP States.
Cell Chem Biol, 24, 2017
|
|
2XST
 
 | | Crystal Structure of the Human Lipocalin 15 | | Descriptor: | 1,2-ETHANEDIOL, LIPOCALIN 15 | | Authors: | Muniz, J.R.C, Gileadi, C, Yue, W.W, Krojer, T, Ugochukwu, E, Phillips, C, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Kavanagh, K.L, Oppermann, U. | | Deposit date: | 2010-09-30 | | Release date: | 2010-10-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Crystal Structure of the Human Lipocalin 15
To be Published
|
|
7BHV
 
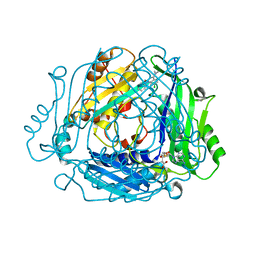 | | Crystal structure of MAT2a bound to allosteric inhibitor and in vivo tool compound 28 | | Descriptor: | 7-chloranyl-4-(dimethylamino)-1-phenyl-quinazolin-2-one, S-ADENOSYLMETHIONINE, S-adenosylmethionine synthase isoform type-2 | | Authors: | Schimpl, M, De Fusco, C, Borjesson, U, Cheung, T, Collie, I, Evans, L, Narasimhan, P, Stubbs, C, Vazquez-Chantada, M, Wagner, D.J, Grondine, M, Tentarelli, S, Underwood, E, Argyrou, A, Bagal, S, Chiarparin, E, Robb, G, Scott, J.S. | | Deposit date: | 2021-01-11 | | Release date: | 2021-04-21 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Fragment-Based Design of a Potent MAT2a Inhibitor and in Vivo Evaluation in an MTAP Null Xenograft Model.
J.Med.Chem., 64, 2021
|
|
7JMF
 
 | | Functional Pathways of Biomolecules Retrieved from Single-particle Snapshots - Frame 42 - State 6 (S6) | | Descriptor: | CALCIUM ION, Peptidyl-prolyl cis-trans isomerase FKBP1B, ZINC ION, ... | | Authors: | Dashti, A, des Georges, A, Frank, J, Ourmazd, A. | | Deposit date: | 2020-07-31 | | Release date: | 2020-08-12 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Retrieving functional pathways of biomolecules from single-particle snapshots.
Nat Commun, 11, 2020
|
|
7BHT
 
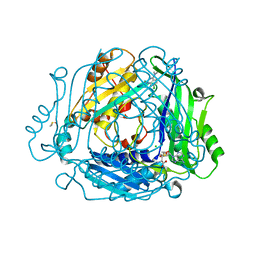 | | Crystal structure of MAT2a with quinazolinone fragment 5 bound in the allosteric site | | Descriptor: | 7-chloranyl-4-(dimethylamino)-1~{H}-quinazolin-2-one, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Schimpl, M, De Fusco, C, Borjesson, U, Cheung, T, Collie, I, Evans, L, Narasimhan, P, Stubbs, C, Vazquez-Chantada, M, Wagner, D.J, Grondine, M, Tentarelli, S, Underwood, E, Argyrou, A, Bagal, S, Chiarparin, E, Robb, G, Scott, J.S. | | Deposit date: | 2021-01-11 | | Release date: | 2021-04-21 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.052 Å) | | Cite: | Fragment-Based Design of a Potent MAT2a Inhibitor and in Vivo Evaluation in an MTAP Null Xenograft Model.
J.Med.Chem., 64, 2021
|
|
6BCC
 
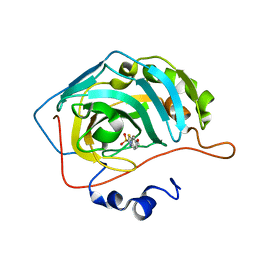 | | Joint X-ray/neutron structure of human carbonic anhydrase II in complex with ethoxzolamide | | Descriptor: | 6-ethoxy-1,3-benzothiazole-2-sulfonamide, Carbonic anhydrase 2, ZINC ION | | Authors: | Kovalevsky, A, McKenna, R, Aggarwal, M. | | Deposit date: | 2017-10-20 | | Release date: | 2018-02-28 | | Last modified: | 2023-10-04 | | Method: | NEUTRON DIFFRACTION (1.8 Å), X-RAY DIFFRACTION | | Cite: | "To Be or Not to Be" Protonated: Atomic Details of Human Carbonic Anhydrase-Clinical Drug Complexes by Neutron Crystallography and Simulation.
Structure, 26, 2018
|
|
7BHX
 
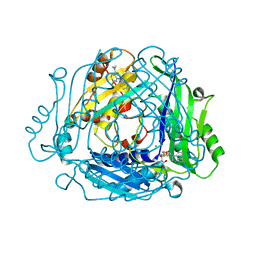 | | Crystal structure of MAT2a bound to allosteric inhibitor (compound 31) | | Descriptor: | 7-chloranyl-4-(dimethylamino)-1-pyridin-3-yl-quinazolin-2-one, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Schimpl, M, De Fusco, C, Borjesson, U, Cheung, T, Collie, I, Evans, L, Narasimhan, P, Stubbs, C, Vazquez-Chantada, M, Wagner, D.J, Grondine, M, Tentarelli, S, Underwood, E, Argyrou, A, Bagal, S, Chiarparin, E, Robb, G, Scott, J.S. | | Deposit date: | 2021-01-11 | | Release date: | 2021-04-21 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Fragment-Based Design of a Potent MAT2a Inhibitor and in Vivo Evaluation in an MTAP Null Xenograft Model.
J.Med.Chem., 64, 2021
|
|
6BDD
 
 | | Crystal structure of Fe(II) unliganded H-NOX protein from K. algicida | | Descriptor: | 2,4-dihydroxyhept-2-ene-1,7-dioic acid aldolase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Bruegger, J.J, Hespen, C.W, Marletta, M.A. | | Deposit date: | 2017-10-22 | | Release date: | 2018-05-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Native Alanine Substitution in the Glycine Hinge Modulates Conformational Flexibility of Heme Nitric Oxide/Oxygen (H-NOX) Sensing Proteins.
ACS Chem. Biol., 13, 2018
|
|
8G4O
 
 | | Native GABA-A receptor from the mouse brain, alpha1-beta2-gamma2 subtype, in complex with didesethylflurazepam and endogenous GABA | | Descriptor: | (5M)-1-(2-aminoethyl)-7-chloro-5-(2-fluorophenyl)-1,3-dihydro-2H-1,4-benzodiazepin-2-one, 2-acetamido-2-deoxy-beta-D-glucopyranose, GAMMA-AMINO-BUTANOIC ACID, ... | | Authors: | Sun, C, Gouaux, E. | | Deposit date: | 2023-02-10 | | Release date: | 2023-09-20 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Cryo-EM structures reveal native GABA A receptor assemblies and pharmacology.
Nature, 622, 2023
|
|
3DYS
 
 | | human phosphodiestrase-5'GMP complex (EP), produced by soaking with 20mM cGMP+20 mM MnCl2+20 mM MgCl2 for 2 hours, and flash-cooled to liquid nitrogen temperature when substrate was still abudant. | | Descriptor: | 3-ISOBUTYL-1-METHYLXANTHINE, FORMIC ACID, GUANOSINE-5'-MONOPHOSPHATE, ... | | Authors: | Liu, S, Mansour, M.N, Dillman, K, Perez, J, Danley, D, Menniti, F. | | Deposit date: | 2008-07-28 | | Release date: | 2008-09-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for the catalytic mechanism of human phosphodiesterase 9.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
8DCD
 
 | | RNA ligase RtcB from Pyrococcus horikoshii in complex with Zn2+ and GTP | | Descriptor: | CHLORIDE ION, GLYCEROL, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Jacewicz, A, Dantuluri, S, Shuman, S. | | Deposit date: | 2022-06-16 | | Release date: | 2022-10-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Structures of RNA ligase RtcB in complexes with divalent cations and GTP.
Rna, 28, 2022
|
|
4L3U
 
 | |
6SOB
 
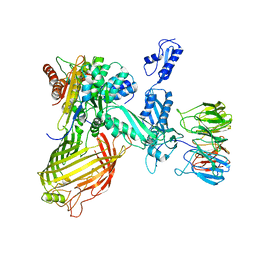 | | BamABCDE in MSP1D1 nanodisc ensemble 1-4 | | Descriptor: | Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamB, Outer membrane protein assembly factor BamC, ... | | Authors: | Iadanza, M.G, Ranson, N.A, Radford, S.E, Higgins, A.J, Calabrese, A.N, Schiffrin, B, White, P. | | Deposit date: | 2019-08-29 | | Release date: | 2020-12-16 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (8.5 Å) | | Cite: | BamABCDE in MSP1D1 nanodisc ensemble 1-4
To Be Published
|
|
9GOO
 
 | |
5CPR
 
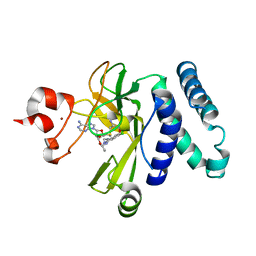 | | The novel SUV4-20 inhibitor A-196 verifies a role for epigenetics in genomic integrity | | Descriptor: | 6,7-dichloro-N-cyclopentyl-4-(pyridin-4-yl)phthalazin-1-amine, Histone-lysine N-methyltransferase SUV420H1, S-ADENOSYLMETHIONINE, ... | | Authors: | Jakob, C.G, Upadhyay, A.K, Sun, C. | | Deposit date: | 2015-07-21 | | Release date: | 2017-01-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | The SUV4-20 inhibitor A-196 verifies a role for epigenetics in genomic integrity.
Nat. Chem. Biol., 13, 2017
|
|
5JUZ
 
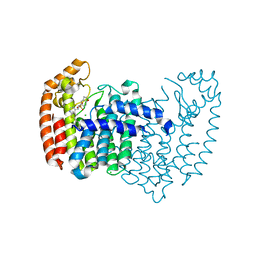 | | Crystal structure of human FPPS in complex with an allosteric inhibitor CL-06-057 | | Descriptor: | CHLORIDE ION, Farnesyl pyrophosphate synthase, [(R)-(2,3-dihydro-1-benzofuran-5-yl){[6-(4-methylphenyl)thieno[2,3-d]pyrimidin-4-yl]amino}methyl]phosphonic acid | | Authors: | Park, J, Magder, A, Leung, C.Y, Tsantrizos, Y.S, Berghuis, A.M. | | Deposit date: | 2016-05-10 | | Release date: | 2017-03-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Pharmacophore Mapping of Thienopyrimidine-Based Monophosphonate (ThP-MP) Inhibitors of the Human Farnesyl Pyrophosphate Synthase.
J. Med. Chem., 60, 2017
|
|
5KA1
 
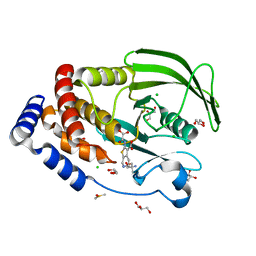 | | Protein Tyrosine Phosphatase 1B Delta helix 7 mutant in complex with TCS401, closed state | | Descriptor: | 2-(OXALYL-AMINO)-4,5,6,7-TETRAHYDRO-THIENO[2,3-C]PYRIDINE-3-CARBOXYLIC ACID, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | Authors: | Choy, M.S, Peti, W, Page, R. | | Deposit date: | 2016-06-01 | | Release date: | 2017-03-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Conformational Rigidity and Protein Dynamics at Distinct Timescales Regulate PTP1B Activity and Allostery.
Mol. Cell, 65, 2017
|
|
