6BRA
 
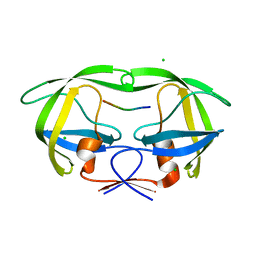 | |
5TYR
 
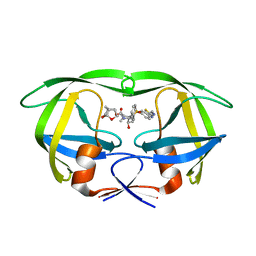 | | X-ray crystal structure of wild type HIV-1 protease in complex with GRL-121 | | Descriptor: | (3S,3aR,5R,7aS,8S)-hexahydro-4H-3,5-methanofuro[2,3-b]pyran-8-yl {(2S,3R)-4-[{[2-(cyclopropylamino)-1,3-benzothiazol-6-yl]sulfonyl}(2-methylpropyl)amino]-3-hydroxy-1-phenylbutan-2-yl}carbamate, Protease | | Authors: | Yedidi, R.S, Hayashi, H, Aoki, M, Das, D, Ghosh, A.K, Mitsuya, H. | | Deposit date: | 2016-11-21 | | Release date: | 2017-10-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A novel central nervous system-penetrating protease inhibitor overcomes human immunodeficiency virus 1 resistance with unprecedented aM to pM potency.
Elife, 6, 2017
|
|
5TQS
 
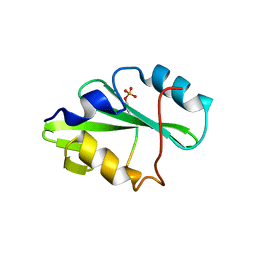 | |
7B94
 
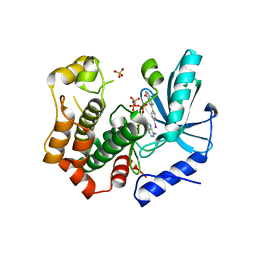 | | MEK1 in complex with compound 6 | | Descriptor: | 2-(4-iodophenyl)-8~{H}-imidazo[1,2-c]pyrimidin-5-one, Dual specificity mitogen-activated protein kinase kinase 1,Dual specificity mitogen-activated protein kinase kinase 1, MAGNESIUM ION, ... | | Authors: | Kack, H, Oster, L. | | Deposit date: | 2020-12-14 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Fragment-Based Discovery of Novel Allosteric MEK1 Binders.
Acs Med.Chem.Lett., 12, 2021
|
|
5YEG
 
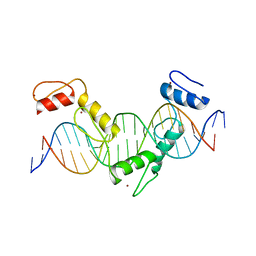 | | Crystal structure of CTCF ZFs4-8-Hs5-1a complex | | Descriptor: | DNA (5'-D(*AP*CP*TP*TP*TP*AP*AP*CP*CP*AP*GP*CP*AP*GP*AP*GP*GP*GP*CP*G)-3'), DNA (5'-D(*TP*CP*GP*CP*CP*CP*TP*CP*TP*GP*CP*TP*GP*GP*TP*TP*AP*AP*AP*G)-3'), Transcriptional repressor CTCF, ... | | Authors: | Yin, M, Wang, J, Wang, M, Li, X. | | Deposit date: | 2017-09-17 | | Release date: | 2017-11-29 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular mechanism of directional CTCF recognition of a diverse range of genomic sites
Cell Res., 27, 2017
|
|
7BE5
 
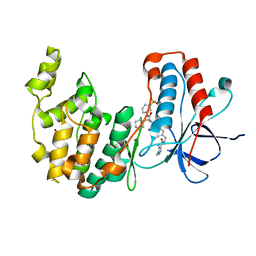 | | Crystal structure of MAP kinase p38 alpha in complex with inhibitor SR276 | | Descriptor: | 1,2-ETHANEDIOL, 5-azanyl-~{N}-[[4-[[(2~{S})-4-cyclohexyl-1-[[(3~{R})-1-methylsulfonylpiperidin-3-yl]amino]-1-oxidanylidene-butan-2-yl]carbamoyl]phenyl]methyl]-1-methyl-pyrazole-4-carboxamide, Mitogen-activated protein kinase 14 | | Authors: | Joerger, A.C, Schroeder, M, Roehm, S, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-12-22 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8000524 Å) | | Cite: | Development of a Selective Dual Discoidin Domain Receptor (DDR)/p38 Kinase Chemical Probe.
J.Med.Chem., 64, 2021
|
|
7BDO
 
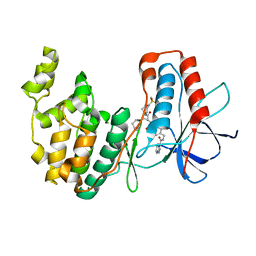 | | MAPK14 bound with SR302 | | Descriptor: | Mitogen-activated protein kinase 14, ~{N}-[[4-[[(2~{S})-4-cyclohexyl-1-[[(3~{S})-1-methylsulfonylpiperidin-3-yl]amino]-1-oxidanylidene-butan-2-yl]carbamoyl]phenyl]methyl]imidazo[1,2-a]pyridine-3-carboxamide | | Authors: | Schroeder, M, Roehm, S, Joerger, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-12-22 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Development of a Selective Dual Discoidin Domain Receptor (DDR)/p38 Kinase Chemical Probe.
J.Med.Chem., 64, 2021
|
|
8F05
 
 | |
4N34
 
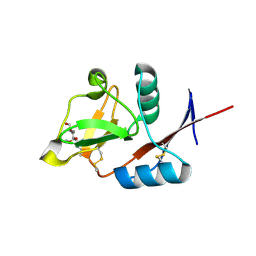 | | Structure of langerin CRD I313 with alpha-MeGlcNAc | | Descriptor: | C-type lectin domain family 4 member K, CALCIUM ION, methyl 2-acetamido-2-deoxy-alpha-D-glucopyranoside | | Authors: | Feinberg, H, Rowntree, T.J.W, Tan, S.L.W, Drickamer, K, Weis, W.I, Taylor, M.E. | | Deposit date: | 2013-10-06 | | Release date: | 2013-11-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Common polymorphisms in human langerin change specificity for glycan ligands.
J.Biol.Chem., 288, 2013
|
|
4N3U
 
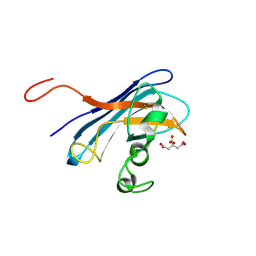 | | Candida albicans Superoxide Dismutase 5 (SOD5), Cu(II) | | Descriptor: | CITRATE ANION, COPPER (II) ION, Potential secreted Cu/Zn superoxide dismutase | | Authors: | Galaleldeen, A, Taylor, A.B, Waninger-Saroni, J.J, Holloway, S.P, Hart, P.J. | | Deposit date: | 2013-10-07 | | Release date: | 2014-04-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Candida albicans SOD5 represents the prototype of an unprecedented class of Cu-only superoxide dismutases required for pathogen defense.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
6CDJ
 
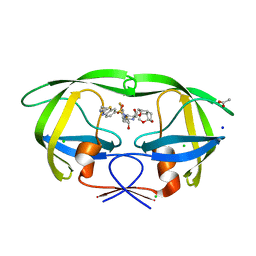 | | HIV-1 wild type protease with GRL-03314A, 6-5-5-ring fused umbrella-like tetrahydropyranofuran as the P2-ligand, a cyclopropylaminobenzothiazole as the P2'-ligand and 3,5-difluorophenylmethyl as the P1-ligand | | Descriptor: | (2aS,4R,4aS,7aS,7bS)-octahydro-2H-1,7-dioxacyclopenta[cd]inden-4-yl [(2S,3R)-4-[{[2-(cyclopropylamino)-1,3-benzothiazol-6-yl]sulfonyl}(2-methylpropyl)amino]-1-(3,5-difluorophenyl)-3-hydroxybutan-2-yl]carbamate, ACETATE ION, CHLORIDE ION, ... | | Authors: | Wang, Y.-F, Agniswamy, J, Weber, I.T. | | Deposit date: | 2018-02-08 | | Release date: | 2018-05-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Design and Synthesis of Highly Potent HIV-1 Protease Inhibitors Containing Tricyclic Fused Ring Systems as Novel P2 Ligands: Structure-Activity Studies, Biological and X-ray Structural Analysis.
J. Med. Chem., 61, 2018
|
|
8EZX
 
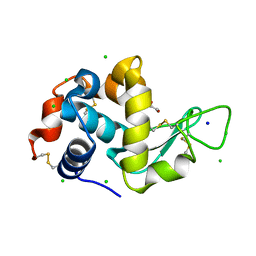 | | Lysozyme Anomalous Dataset at 293 K and 7.1 keV | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysozyme C, ... | | Authors: | Doukov, T, Yabukarski, F, Herschlag, D. | | Deposit date: | 2022-11-01 | | Release date: | 2023-03-15 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Obtaining anomalous and ensemble information from protein crystals from 220 K up to physiological temperatures.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
7KVP
 
 | | Human CYP3A4 bound to an inhibitor | | Descriptor: | Cytochrome P450 3A4, PROTOPORPHYRIN IX CONTAINING FE, tert-butyl [(2R)-1-(naphthalen-1-yl)-3-{[(2S)-1-oxo-3-phenyl-1-{[3-(pyridin-3-yl)propyl]amino}propan-2-yl]sulfanyl}propan-2-yl]carbamate | | Authors: | Sevrioukova, I. | | Deposit date: | 2020-11-28 | | Release date: | 2021-01-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Rational Design of CYP3A4 Inhibitors: A One-Atom Linker Elongation in Ritonavir-Like Compounds Leads to a Marked Improvement in the Binding Strength.
Int J Mol Sci, 22, 2021
|
|
5YOJ
 
 | | Structure of A17 HIV-1 Protease in Complex with Inhibitor KNI-1657 | | Descriptor: | (4R)-N-[(2,6-dimethylphenyl)methyl]-3-[(2S,3S)-3-[[(2S)-2-[(7-methoxy-1-benzofuran-2-yl)carbonylamino]-2-[(3R)-oxolan-3 -yl]ethanoyl]amino]-2-oxidanyl-4-phenyl-butanoyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxamide, A17 HIV-1 protease, GLYCEROL | | Authors: | Adachi, M, Hidaka, K, Kuroki, R, Kiso, Y. | | Deposit date: | 2017-10-29 | | Release date: | 2018-07-11 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Identification of Highly Potent Human Immunodeficiency Virus Type-1 Protease Inhibitors against Lopinavir and Darunavir Resistant Viruses from Allophenylnorstatine-Based Peptidomimetics with P2 Tetrahydrofuranylglycine.
J. Med. Chem., 61, 2018
|
|
4MLW
 
 | |
8EK1
 
 | |
6CPB
 
 | |
5YIN
 
 | |
6IOM
 
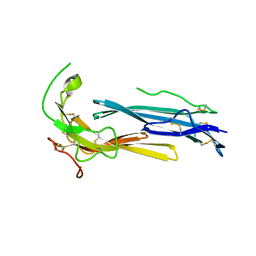 | | Crystal structure of human C4.4A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Ly6/PLAUR domain-containing protein 3 | | Authors: | Huang, M.D, Jiang, Y.B, Yuan, C, Lin, L. | | Deposit date: | 2018-10-30 | | Release date: | 2020-01-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.594 Å) | | Cite: | Crystal Structures of Human C4.4A Reveal the Unique Association of Ly6/uPAR/alpha-neurotoxin Domain
Int J Biol Sci, 16, 2020
|
|
8EZO
 
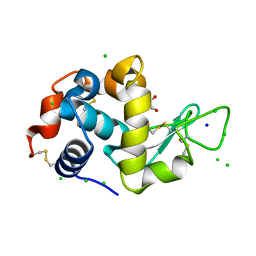 | | Lysozyme Anomalous Dataset at 220 K and 7.1 keV | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysozyme C, ... | | Authors: | Doukov, T, Yabukarski, F. | | Deposit date: | 2022-11-01 | | Release date: | 2023-03-15 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Obtaining anomalous and ensemble information from protein crystals from 220 K up to physiological temperatures.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
7L3U
 
 | |
6IP9
 
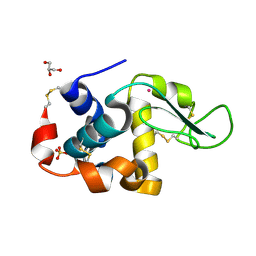 | | Crystal Structure of Lanthanum ion (La3+) bound bovine alpha-lactalbumin | | Descriptor: | Alpha-lactalbumin, GLYCEROL, LANTHANUM (III) ION, ... | | Authors: | Prakash, P, Yarramala, S.D, Rao, C.P, Bhaumik, P. | | Deposit date: | 2018-11-02 | | Release date: | 2019-02-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Cytotoxicity of apo bovine alpha-lactalbumin complexed with La3+on cancer cells supported by its high resolution crystal structure.
Sci Rep, 9, 2019
|
|
8F0B
 
 | | Lysozyme Anomalous Dataset at 240 K and 7.1 keV | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysozyme C, ... | | Authors: | Doukov, T, Yabukarski, F, Herschlag, D. | | Deposit date: | 2022-11-02 | | Release date: | 2023-03-15 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Obtaining anomalous and ensemble information from protein crystals from 220 K up to physiological temperatures.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
7L61
 
 | |
7L67
 
 | |
