6KSD
 
 | |
6KSC
 
 | |
8IJ1
 
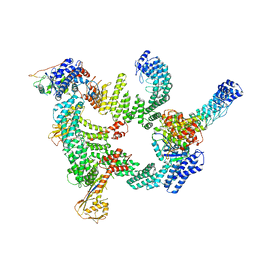 | | Protomer 1 and 2 of the asymmetry trimer of the Cul2-Rbx1-EloBC-FEM1B ubiquitin ligase complex | | Descriptor: | Cullin-2, E3 ubiquitin-protein ligase RBX1, Elongin-B, ... | | Authors: | Dai, Z, Liang, L, Yin, Y.X. | | Deposit date: | 2023-02-24 | | Release date: | 2024-02-28 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structural insights into the ubiquitylation strategy of the oligomeric CRL2 FEM1B E3 ubiquitin ligase.
Embo J., 43, 2024
|
|
3Q4L
 
 | | Structure of a small peptide ligand bound to E.coli DNA sliding clamp | | Descriptor: | DNA polymerase III subunit beta, SODIUM ION, peptide ligand | | Authors: | Wolff, P, Olieric, V, Briand, J.P, Chaloin, O, Dejaegere, A, Dumas, P, Ennifar, E, Guichard, G, Wagner, J, Burnouf, D. | | Deposit date: | 2010-12-23 | | Release date: | 2011-12-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure-based design of short peptide ligands binding onto the E. coli processivity ring.
J.Med.Chem., 54, 2011
|
|
8KHP
 
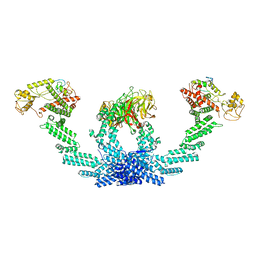 | | CULLIN3-KLHL22-RBX1 E3 ligase | | Descriptor: | Cullin-3, E3 ubiquitin-protein ligase RBX1, Kelch-like protein 22 | | Authors: | Su, M.-Y, Su, M.-Y. | | Deposit date: | 2023-08-22 | | Release date: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (3.67 Å) | | Cite: | Cryo-EM structure of the KLHL22 E3 ligase bound to an oligomeric metabolic enzyme.
Structure, 31, 2023
|
|
9J8E
 
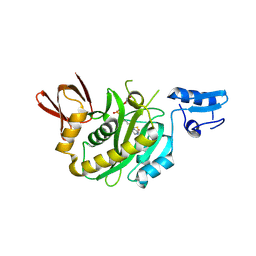 | | Structural insights into BirA from Haemophilus influenzae, a bifunctional protein as a biotin protein ligase and a transcriptional repressor | | Descriptor: | BIOTINYL-5-AMP, Bifunctional ligase/repressor BirA | | Authors: | Lee, J.Y, Jeong, K.H, Son, S.B, Ko, J.H. | | Deposit date: | 2024-08-21 | | Release date: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural insights into BirA from Haemophilus influenzae, a bifunctional protein as a biotin protein ligase and a transcriptional repressor.
Biochem.Biophys.Res.Commun., 733, 2024
|
|
4I1H
 
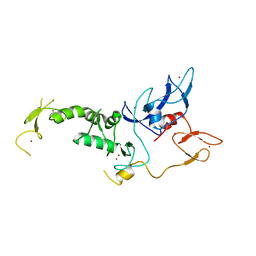 | | Structure of Parkin E3 ligase | | Descriptor: | E3 ubiquitin-protein ligase parkin, ZINC ION | | Authors: | Lougheed, J.C, Brecht, E, Yao, N.H. | | Deposit date: | 2012-11-20 | | Release date: | 2013-06-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and function of Parkin E3 ubiquitin ligase reveals aspects of RING and HECT ligases.
Nat Commun, 4, 2013
|
|
1S68
 
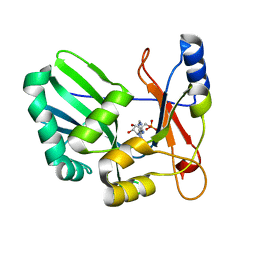 | | Structure and Mechanism of RNA Ligase | | Descriptor: | ADENOSINE MONOPHOSPHATE, RNA Ligase 2 | | Authors: | Ho, C.K, Wang, L.K, Lima, C.D, Shuman, S. | | Deposit date: | 2004-01-22 | | Release date: | 2004-02-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and mechanism of RNA ligase.
Structure, 12, 2004
|
|
4C5A
 
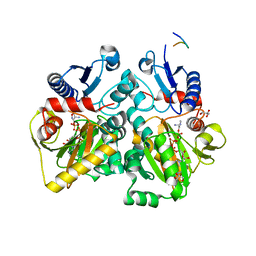 | | The X-ray crystal structures of D-alanyl-D-alanine ligase in complex ADP and D-cycloserine phosphate | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, D-ALANINE--D-ALANINE LIGASE, GLYCEROL, ... | | Authors: | Batson, S, Majce, V, Lloyd, A.J, Rea, D, Fishwick, C.W.G, Simmons, K.J, Fulop, V, Roper, D.I. | | Deposit date: | 2013-09-10 | | Release date: | 2015-01-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Inhibition of D-Ala:D-Ala ligase through a phosphorylated form of the antibiotic D-cycloserine.
Nat Commun, 8, 2017
|
|
8JE1
 
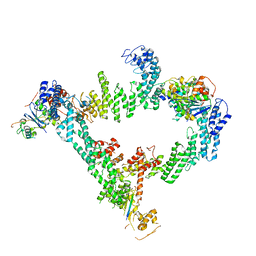 | | An asymmetry dimer of the Cul2-Rbx1-EloBC-FEM1B ubiquitin ligase complexed with BEX2 | | Descriptor: | Cullin-2, E3 ubiquitin-protein ligase RBX1, Elongin-B, ... | | Authors: | Dai, Z, Liang, L, Yin, Y.X. | | Deposit date: | 2023-05-15 | | Release date: | 2024-02-28 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (3.95 Å) | | Cite: | Structural insights into the ubiquitylation strategy of the oligomeric CRL2 FEM1B E3 ubiquitin ligase.
Embo J., 43, 2024
|
|
5O6C
 
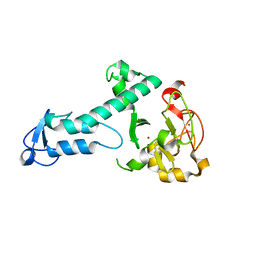 | | Crystal Structure of a threonine-selective RCR E3 ligase | | Descriptor: | E3 ubiquitin-protein ligase MYCBP2, ZINC ION | | Authors: | Pao, K.-C, Rafie, K.Z, van Aalten, D, Virdee, S. | | Deposit date: | 2017-06-06 | | Release date: | 2018-04-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Activity-based E3 ligase profiling uncovers an E3 ligase with esterification activity.
Nature, 556, 2018
|
|
4C5B
 
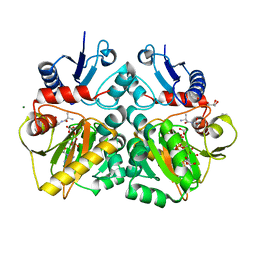 | | The X-ray crystal structure of D-alanyl-D-alanine ligase in complex with ATP and D-ala-D-ala | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CARBONATE ION, D-ALANINE, ... | | Authors: | Batson, S, Majce, V, Lloyd, A.J, Rea, D, Fishwick, C.W.G, Simmons, K.J, Fulop, V, Roper, D.I. | | Deposit date: | 2013-09-10 | | Release date: | 2015-01-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Inhibition of D-Ala:D-Ala ligase through a phosphorylated form of the antibiotic D-cycloserine.
Nat Commun, 8, 2017
|
|
4C5C
 
 | | The X-ray crystal structure of D-alanyl-D-alanine ligase in complex with ADP and D-ala-D-ala | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, D-ALANINE, D-ALANINE--D-ALANINE LIGASE, ... | | Authors: | Batson, S, Majce, V, Lloyd, A.J, Rea, D, Fishwick, C.W.G, Simmons, K.J, Fulop, V, Roper, D.I. | | Deposit date: | 2013-09-10 | | Release date: | 2015-01-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Inhibition of D-Ala:D-Ala ligase through a phosphorylated form of the antibiotic D-cycloserine.
Nat Commun, 8, 2017
|
|
2OOB
 
 | |
4XOM
 
 | | Coenzyme F420:L-glutamate ligase (FbiB) from Mycobacterium tuberculosis (C-terminal domain). | | Descriptor: | Coenzyme F420:L-glutamate ligase, SULFATE ION | | Authors: | Rehan, A.M, Bashiri, G, Baker, H.M, Baker, E.N, Squire, C.J. | | Deposit date: | 2015-01-16 | | Release date: | 2016-02-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Elongation of the Poly-gamma-glutamate Tail of F420 Requires Both Domains of the F420: gamma-Glutamyl Ligase (FbiB) of Mycobacterium tuberculosis.
J.Biol.Chem., 291, 2016
|
|
2C5U
 
 | | T4 RNA Ligase (Rnl1) Crystal Structure | | Descriptor: | CALCIUM ION, CHLORIDE ION, DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, ... | | Authors: | El Omari, K, Ren, J, Bird, L.E, Bona, M.K, Klarmann, G, LeGrice, S.F.J, Stammers, D.K. | | Deposit date: | 2005-11-01 | | Release date: | 2005-11-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Molecular Architecture and Ligand Recognition Determinants for T4 RNA Ligase
J.Biol.Chem., 281, 2006
|
|
1DGS
 
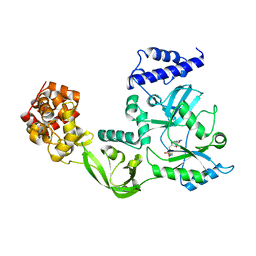 | | CRYSTAL STRUCTURE OF NAD+-DEPENDENT DNA LIGASE FROM T. FILIFORMIS | | Descriptor: | ADENOSINE MONOPHOSPHATE, DNA LIGASE, ZINC ION | | Authors: | Lee, J.Y, Chang, C, Song, H.K, Kwon, S.T, Suh, S.W. | | Deposit date: | 1999-11-25 | | Release date: | 2000-11-27 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of NAD(+)-dependent DNA ligase: modular architecture and functional implications.
EMBO J., 19, 2000
|
|
6J1Y
 
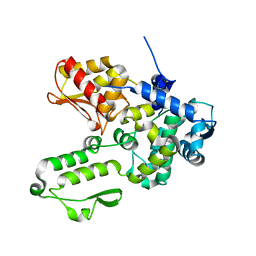 | | Semi-open conformation E3 ligase | | Descriptor: | GLYCEROL, NEDD4-like E3 ubiquitin-protein ligase WWP1 | | Authors: | Liu, Z.H. | | Deposit date: | 2018-12-30 | | Release date: | 2019-07-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | A multi-lock inhibitory mechanism for fine-tuning enzyme activities of the HECT family E3 ligases.
Nat Commun, 10, 2019
|
|
8E0Q
 
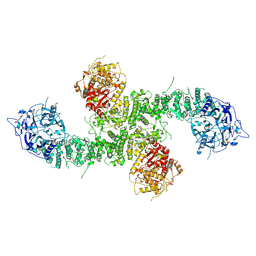 | | Structure of the human UBR5 HECT-type E3 ubiquitin ligase in a C2 symmetric dimeric form | | Descriptor: | E3 ubiquitin-protein ligase UBR5, ZINC ION | | Authors: | Wang, F, He, Q, Lin, G, Li, H. | | Deposit date: | 2022-08-09 | | Release date: | 2023-04-19 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (2.66 Å) | | Cite: | Structure of the human UBR5 E3 ubiquitin ligase.
Structure, 31, 2023
|
|
4R9M
 
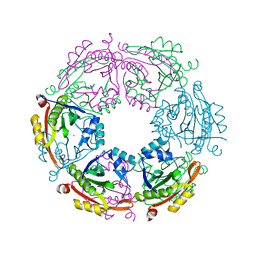 | | Crystal structure of spermidine N-acetyltransferase from Escherichia coli | | Descriptor: | MAGNESIUM ION, Spermidine N(1)-acetyltransferase | | Authors: | Filippova, E.V, Minasov, G, Kiryukhina, O, Shuvalova, L, Grimshaw, S, Wolfe, A.J, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-09-05 | | Release date: | 2014-11-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Analysis of crystalline and solution states of ligand-free spermidine N-acetyltransferase (SpeG) from Escherichia coli.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
6GMO
 
 | |
8EWI
 
 | | Structure of the human UBR5 HECT-type E3 ubiquitin ligase in a tetrameric form | | Descriptor: | E3 ubiquitin-protein ligase UBR5, ZINC ION | | Authors: | Wang, F, He, Q, Lin, G, Li, H. | | Deposit date: | 2022-10-23 | | Release date: | 2023-04-19 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of the human UBR5 E3 ubiquitin ligase.
Structure, 31, 2023
|
|
3OEM
 
 | |
7P3B
 
 | | Human RNA ligase RTCB in complex with GMP and Co(II) | | Descriptor: | ACETATE ION, COBALT (II) ION, FORMIC ACID, ... | | Authors: | Kroupova, A, Ackle, F, Jinek, M. | | Deposit date: | 2021-07-07 | | Release date: | 2021-12-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular architecture of the human tRNA ligase complex.
Elife, 10, 2021
|
|
4RFW
 
 | | Crystal structure of human retinoid X Receptor alpha-ligand binding domain complex with 9cUAB70 and the coactivator peptide GRIP-1 | | Descriptor: | (2E,4E,6Z,8E)-3,7-dimethyl-8-(6,7,8,9-tetrahydro-5H-benzo[7]annulen-5-ylidene)octa-2,4,6-trienoic acid, Nuclear receptor coactivator 2, Retinoic acid receptor RXR-alpha | | Authors: | Xia, G, Muccio, D.D, Smith, C.D. | | Deposit date: | 2014-09-28 | | Release date: | 2015-09-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Conformationally Defined Rexinoids and Their Efficacy in the Prevention of Mammary Cancers.
J.Med.Chem., 58, 2015
|
|
