5KIT
 
 | |
1QHS
 
 | |
8J40
 
 | | Crystal Structure of CATB8 in complex with chloramphenicol | | Descriptor: | CHLORAMPHENICOL, CatB8, GLYCEROL, ... | | Authors: | Liao, J, Kuang, L, Jiang, Y. | | Deposit date: | 2023-04-18 | | Release date: | 2024-02-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Chloramphenicol Binding Sites of Acinetobacter baumannii Chloramphenicol Acetyltransferase CatB8.
Acs Infect Dis., 10, 2024
|
|
4YB6
 
 | | Adenosine triphosphate phosphoribosyltransferase from Campylobacter jejuni in complex with the inhibitors AMP and histidine | | Descriptor: | ADENOSINE MONOPHOSPHATE, ATP phosphoribosyltransferase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Mittelstaedt, G, Moggre, G.-J, Parker, E.J. | | Deposit date: | 2015-02-18 | | Release date: | 2016-03-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Campylobacter jejuni adenosine triphosphate phosphoribosyltransferase is an active hexamer that is allosterically controlled by the twisting of a regulatory tail.
Protein Sci., 25, 2016
|
|
4D3H
 
 | | Structure of PstA | | Descriptor: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, PSTA | | Authors: | Campeotto, I, Freemont, P, Grundling, A. | | Deposit date: | 2014-10-22 | | Release date: | 2014-12-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Complex Structure and Biochemical Characterization of the Staphylococcus Aureus Cyclic Di-AMP Binding Protein Psta, the Founding Member of a New Signal Transduction Protein Family
J.Biol.Chem., 290, 2015
|
|
4D3G
 
 | | Structure of PstA | | Descriptor: | PSTA | | Authors: | Campeotto, I, Freemont, P, Grundling, A. | | Deposit date: | 2014-10-22 | | Release date: | 2014-12-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Complex Structure and Biochemical Characterization of the Staphylococcus Aureus Cyclic Di-AMP Binding Protein Psta, the Founding Member of a New Signal Transduction Protein Family
J.Biol.Chem., 290, 2015
|
|
1Q1K
 
 | | Structure of ATP-phosphoribosyltransferase from E. coli complexed with PR-ATP | | Descriptor: | ATP phosphoribosyltransferase, L(+)-TARTARIC ACID, PHOSPHORIBOSYL ATP | | Authors: | Lohkamp, B, McDermott, G, Coggins, J.R, Lapthorn, A.J. | | Deposit date: | 2003-07-21 | | Release date: | 2004-03-02 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The structure of Escherichia coli ATP-phosphoribosyltransferase: identification of substrate binding sites and mode of AMP inhibition
J.Mol.Biol., 336, 2004
|
|
8HUB
 
 | | AMP deaminase 2 in complex with an inhibitor | | Descriptor: | 3,3-dimethyl-4-(phenylmethyl)-2~{H}-quinoxaline-1-carboxamide, AMP deaminase 2, ZINC ION | | Authors: | Adachi, T, Doi, S. | | Deposit date: | 2022-12-23 | | Release date: | 2023-01-18 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | The discovery of 3,3-dimethyl-1,2,3,4-tetrahydroquinoxaline-1-carboxamides as AMPD2 inhibitors with a novel mechanism of action.
Bioorg.Med.Chem.Lett., 80, 2023
|
|
5FBU
 
 | | Crystal structure of rifampin phosphotransferase RPH-Lm from Listeria monocytogenes in complex with rifampin-phosphate | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, Phosphoenolpyruvate synthase, ... | | Authors: | Stogios, P.J, Wawrzak, Z, Skarina, T, Yim, V, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-12-14 | | Release date: | 2015-12-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Rifampin phosphotransferase is an unusual antibiotic resistance kinase.
Nat Commun, 7, 2016
|
|
5FBT
 
 | | Crystal structure of rifampin phosphotransferase RPH-Lm from Listeria monocytogenes in complex with rifampin | | Descriptor: | CHLORIDE ION, Phosphoenolpyruvate synthase, Rifampin | | Authors: | Stogios, P.J, Wawrzak, Z, Skarina, T, Yim, V, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-12-14 | | Release date: | 2015-12-30 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.702 Å) | | Cite: | Rifampin phosphotransferase is an unusual antibiotic resistance kinase.
Nat Commun, 7, 2016
|
|
4UM4
 
 | |
4WQ6
 
 | | The crystal structure of human Nicotinamide phosphoribosyltransferase (NAMPT) in complex with N-(4-{(S)-[1-(2-methylpropyl)piperidin-4-yl]sulfinyl}benzyl)furo[2,3-c]pyridine-2-carboxamide inhibitor (compound 21) | | Descriptor: | 1,2-ETHANEDIOL, N-(4-{(S)-[1-(2-methylpropyl)piperidin-4-yl]sulfinyl}benzyl)furo[2,3-c]pyridine-2-carboxamide, Nicotinamide phosphoribosyltransferase, ... | | Authors: | Li, D, Wang, W. | | Deposit date: | 2014-10-21 | | Release date: | 2015-02-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Identification of nicotinamide phosphoribosyltransferase (NAMPT) inhibitors with no evidence of CYP3A4 time-dependent inhibition and improved aqueous solubility.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
1FT7
 
 | | AAP COMPLEXED WITH L-LEUCINEPHOSPHONIC ACID | | Descriptor: | BACTERIAL LEUCYL AMINOPEPTIDASE, LEUCINE PHOSPHONIC ACID, POTASSIUM ION, ... | | Authors: | Stamper, C, Bennett, B, Holz, R, Petsko, G, Ringe, D. | | Deposit date: | 2000-09-11 | | Release date: | 2000-10-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Inhibition of the aminopeptidase from Aeromonas proteolytica by L-leucinephosphonic acid. Spectroscopic and crystallographic characterization of the transition state of peptide hydrolysis.
Biochemistry, 40, 2001
|
|
5L1H
 
 | | AMPA subtype ionotropic glutamate receptor GluA2 in complex with noncompetitive inhibitor GYKI53655 | | Descriptor: | (8R)-5-(4-aminophenyl)-N,8-dimethyl-8,9-dihydro-2H,7H-[1,3]dioxolo[4,5-h][2,3]benzodiazepine-7-carboxamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, Glutamate receptor 2 | | Authors: | Yelshanskaya, M.V, Singh, A.K, Sampson, J.M, Sobolevsky, A.I. | | Deposit date: | 2016-07-29 | | Release date: | 2016-10-19 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (3.801 Å) | | Cite: | Structural Bases of Noncompetitive Inhibition of AMPA-Subtype Ionotropic Glutamate Receptors by Antiepileptic Drugs.
Neuron, 91, 2016
|
|
5L1B
 
 | | AMPA subtype ionotropic glutamate receptor GluA2 in Apo state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Glutamate receptor 2,Glutamate receptor 2 | | Authors: | Yelshanskaya, M.V, Singh, A.K, Sampson, J.M, Sobolevsky, A.I. | | Deposit date: | 2016-07-28 | | Release date: | 2016-10-19 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Structural Bases of Noncompetitive Inhibition of AMPA-Subtype Ionotropic Glutamate Receptors by Antiepileptic Drugs.
Neuron, 91, 2016
|
|
5L1F
 
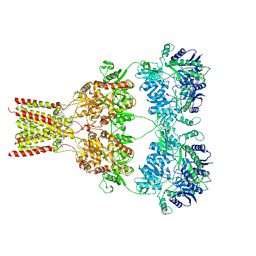 | | AMPA subtype ionotropic glutamate receptor GluA2 in complex with noncompetitive inhibitor Perampanel | | Descriptor: | 2-(6'-oxo-1'-phenyl[1',6'-dihydro[2,3'-bipyridine]]-5'-yl)benzonitrile, 2-acetamido-2-deoxy-beta-D-glucopyranose, Glutamate receptor 2 | | Authors: | Yelshanskaya, M.V, Singh, A.K, Sampson, J.M, Sobolevsky, A.I. | | Deposit date: | 2016-07-29 | | Release date: | 2016-10-19 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Structural Bases of Noncompetitive Inhibition of AMPA-Subtype Ionotropic Glutamate Receptors by Antiepileptic Drugs.
Neuron, 91, 2016
|
|
5L1E
 
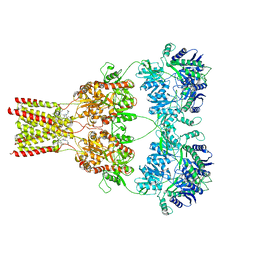 | | AMPA subtype ionotropic glutamate receptor GluA2 in complex with noncompetitive inhibitor CP465022 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 3-(2-chlorophenyl)-2-(2-{6-[(diethylamino)methyl]pyridin-2-yl}ethyl)-6-fluoroquinazolin-4(3H)-one, Glutamate receptor 2 | | Authors: | Yelshanskaya, M.V, Singh, A.K, Sampson, J.M, Sobolevsky, A.I. | | Deposit date: | 2016-07-29 | | Release date: | 2016-10-19 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (4.37 Å) | | Cite: | Structural Bases of Noncompetitive Inhibition of AMPA-Subtype Ionotropic Glutamate Receptors by Antiepileptic Drugs.
Neuron, 91, 2016
|
|
6VVV
 
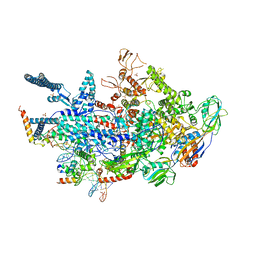 | | Crystal structure of a Mycobacterium smegmatis transcription initiation complex with Rifampicin-resistant RNA polymerase | | Descriptor: | DNA (26-MER), DNA (31-MER), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Lilic, M, Darst, S.A, Campbell, E.A. | | Deposit date: | 2020-02-18 | | Release date: | 2020-10-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The antibiotic sorangicin A inhibits promoter DNA unwinding in a Mycobacterium tuberculosis rifampicin-resistant RNA polymerase.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6VVT
 
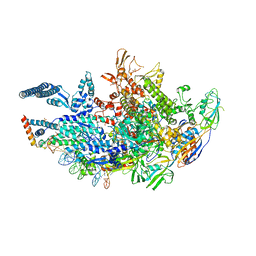 | | Crystal structure of a Mycobacterium smegmatis transcription initiation complex with Rifampicin-resistant RNA polymerase and antibiotic Sorangicin | | Descriptor: | 1,2-ETHANEDIOL, DNA (26-MER), DNA (31-MER), ... | | Authors: | Lilic, M, Darst, S.A, Campbell, E.A. | | Deposit date: | 2020-02-18 | | Release date: | 2020-10-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.901 Å) | | Cite: | The antibiotic sorangicin A inhibits promoter DNA unwinding in a Mycobacterium tuberculosis rifampicin-resistant RNA polymerase.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
2BLS
 
 | |
3BLS
 
 | | AMPC BETA-LACTAMASE FROM ESCHERICHIA COLI | | Descriptor: | AMPC BETA-LACTAMASE, M-AMINOPHENYLBORONIC ACID | | Authors: | Usher, K.C, Shoichet, B.K, Remington, S.J. | | Deposit date: | 1998-06-04 | | Release date: | 1998-08-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Three-dimensional structure of AmpC beta-lactamase from Escherichia coli bound to a transition-state analogue: possible implications for the oxyanion hypothesis and for inhibitor design.
Biochemistry, 37, 1998
|
|
1J3G
 
 | | Solution structure of Citrobacter Freundii AmpD | | Descriptor: | AmpD protein, ZINC ION | | Authors: | Liepinsh, E, Genereux, C, Dehareng, D, Joris, B, Otting, G. | | Deposit date: | 2003-01-31 | | Release date: | 2003-02-18 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of Citrobacter freundii AmpD, Comparison with Bacteriophage T7 Lysozyme
and Homology with PGRP Domains
J.Mol.Biol., 327, 2003
|
|
5L1G
 
 | | AMPA subtype ionotropic glutamate receptor GluA2 in complex with GYKI-Br | | Descriptor: | (8R)-5-(4-amino-3-bromophenyl)-N,8-dimethyl-8,9-dihydro-2H,7H-[1,3]dioxolo[4,5-h][2,3]benzodiazepine-7-carboxamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, Glutamate receptor 2 | | Authors: | Yelshanskaya, M.V, Singh, A.K, Sampson, J.M, Sobolevsky, A.I. | | Deposit date: | 2016-07-29 | | Release date: | 2016-10-19 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (4.507 Å) | | Cite: | Structural Bases of Noncompetitive Inhibition of AMPA-Subtype Ionotropic Glutamate Receptors by Antiepileptic Drugs.
Neuron, 91, 2016
|
|
4O16
 
 | | Structural Basis for Resistance to Diverse Classes of NAMPT Inhibitors | | Descriptor: | 1,2-ETHANEDIOL, 6-({4-[(3,5-difluorophenyl)sulfonyl]benzyl}carbamoyl)-1-(5-O-phosphono-beta-D-ribofuranosyl)imidazo[1,2-a]pyridin-1-ium, Nicotinamide phosphoribosyltransferase, ... | | Authors: | Oh, A, Coons, M, Brillantes, B, Wang, W. | | Deposit date: | 2013-12-15 | | Release date: | 2014-10-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.783 Å) | | Cite: | Structural Basis for Resistance to Diverse Classes of NAMPT Inhibitors.
Plos One, 9, 2014
|
|
4O19
 
 | | The crystal structure of a mutant NAMPT (G217V) | | Descriptor: | 1,2-ETHANEDIOL, Nicotinamide phosphoribosyltransferase, PHOSPHATE ION | | Authors: | Oh, A, Coons, M, Brillantes, B, Wang, W. | | Deposit date: | 2013-12-15 | | Release date: | 2014-10-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Basis for Resistance to Diverse Classes of NAMPT Inhibitors.
Plos One, 9, 2014
|
|
