2R3G
 
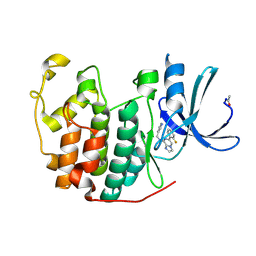 | | Crystal Structure of Cyclin-Dependent Kinase 2 with inhibitor | | Descriptor: | 6-(2-fluorophenyl)-N-(pyridin-3-ylmethyl)imidazo[1,2-a]pyrazin-8-amine, Cell division protein kinase 2 | | Authors: | Fischmann, T.O, Hruza, A.W, Madison, V.M, Duca, J.S. | | Deposit date: | 2007-08-29 | | Release date: | 2008-01-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure-guided discovery of cyclin-dependent kinase inhibitors.
Biopolymers, 89, 2008
|
|
2R3H
 
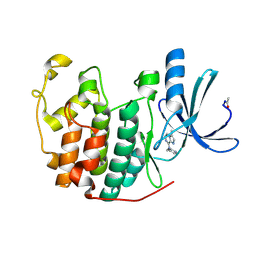 | | Crystal Structure of Cyclin-Dependent Kinase 2 with inhibitor | | Descriptor: | 3-methyl-N-(pyridin-4-ylmethyl)imidazo[1,2-a]pyrazin-8-amine, Cell division protein kinase 2 | | Authors: | Fischmann, T.O, Hruza, A.W, Madison, V.M, Duca, J.S. | | Deposit date: | 2007-08-29 | | Release date: | 2008-01-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure-guided discovery of cyclin-dependent kinase inhibitors.
Biopolymers, 89, 2008
|
|
2R3I
 
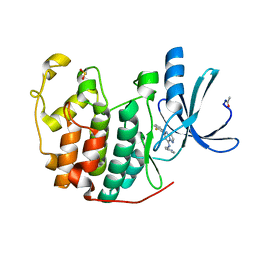 | | Crystal Structure of Cyclin-Dependent Kinase 2 with inhibitor | | Descriptor: | 5-(2-fluorophenyl)-N-(pyridin-4-ylmethyl)pyrazolo[1,5-a]pyrimidin-7-amine, Cell division protein kinase 2 | | Authors: | Fischmann, T.O, Hruza, A.W, Madison, V.M, Duca, J.S. | | Deposit date: | 2007-08-29 | | Release date: | 2008-01-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Structure-guided discovery of cyclin-dependent kinase inhibitors.
Biopolymers, 89, 2008
|
|
2R3J
 
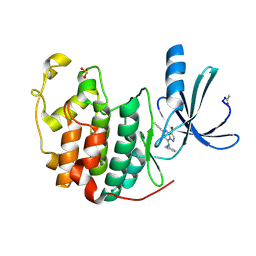 | | Crystal Structure of Cyclin-Dependent Kinase 2 with inhibitor | | Descriptor: | 3-bromo-5-phenyl-N-(pyridin-3-ylmethyl)pyrazolo[1,5-a]pyrimidin-7-amine, Cell division protein kinase 2 | | Authors: | Fischmann, T.O, Hruza, A.W, Madison, V.M, Duca, J.S. | | Deposit date: | 2007-08-29 | | Release date: | 2008-01-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure-guided discovery of cyclin-dependent kinase inhibitors.
Biopolymers, 89, 2008
|
|
2R3K
 
 | | Crystal Structure of Cyclin-Dependent Kinase 2 with inhibitor | | Descriptor: | 3-bromo-5-phenyl-N-(pyrimidin-5-ylmethyl)pyrazolo[1,5-a]pyridin-7-amine, Cell division protein kinase 2 | | Authors: | Fischmann, T.O, Hruza, A.W, Madison, V.M, Duca, J.S. | | Deposit date: | 2007-08-29 | | Release date: | 2008-01-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure-guided discovery of cyclin-dependent kinase inhibitors.
Biopolymers, 89, 2008
|
|
2R3L
 
 | | Crystal Structure of Cyclin-Dependent Kinase 2 with inhibitor | | Descriptor: | 3-bromo-6-phenyl-N-(pyrimidin-5-ylmethyl)imidazo[1,2-a]pyridin-8-amine, Cell division protein kinase 2 | | Authors: | Fischmann, T.O, Hruza, A.W, Madison, V.M, Duca, J.S. | | Deposit date: | 2007-08-29 | | Release date: | 2008-01-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure-guided discovery of cyclin-dependent kinase inhibitors.
Biopolymers, 89, 2008
|
|
2R3M
 
 | | Crystal Structure of Cyclin-Dependent Kinase 2 with inhibitor | | Descriptor: | Cell division protein kinase 2, N-((2-aminopyrimidin-5-yl)methyl)-5-(2,6-difluorophenyl)-3-ethylpyrazolo[1,5-a]pyrimidin-7-amine | | Authors: | Fischmann, T.O, Hruza, A.W, Madison, V.M, Duca, J.S. | | Deposit date: | 2007-08-29 | | Release date: | 2008-01-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure-guided discovery of cyclin-dependent kinase inhibitors.
Biopolymers, 89, 2008
|
|
2R3N
 
 | | Crystal Structure of Cyclin-Dependent Kinase 2 with inhibitor | | Descriptor: | 3-cyclopropyl-5-phenyl-N-(pyridin-3-ylmethyl)pyrazolo[1,5-a]pyrimidin-7-amine, Cell division protein kinase 2 | | Authors: | Fischmann, T.O, Hruza, A.W, Madison, V.M, Duca, J.S. | | Deposit date: | 2007-08-29 | | Release date: | 2008-01-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Structure-guided discovery of cyclin-dependent kinase inhibitors.
Biopolymers, 89, 2008
|
|
2R3O
 
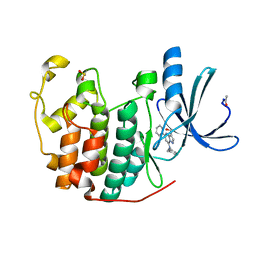 | | Crystal Structure of Cyclin-Dependent Kinase 2 with inhibitor | | Descriptor: | (5-phenyl-7-(pyridin-3-ylmethylamino)pyrazolo[1,5-a]pyrimidin-3-yl)methanol, Cell division protein kinase 2 | | Authors: | Fischmann, T.O, Hruza, A.W, Madison, V.M, Duca, J.S. | | Deposit date: | 2007-08-29 | | Release date: | 2008-01-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-guided discovery of cyclin-dependent kinase inhibitors.
Biopolymers, 89, 2008
|
|
2R3P
 
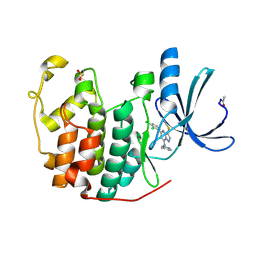 | | Crystal Structure of Cyclin-Dependent Kinase 2 with inhibitor | | Descriptor: | 5-(2,3-dichlorophenyl)-N-(pyridin-4-ylmethyl)-3-thiocyanatopyrazolo[1,5-a]pyrimidin-7-amine, Cell division protein kinase 2 | | Authors: | Fischmann, T.O, Hruza, A.W, Madison, V.M, Duca, J.S. | | Deposit date: | 2007-08-29 | | Release date: | 2008-01-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Structure-guided discovery of cyclin-dependent kinase inhibitors.
Biopolymers, 89, 2008
|
|
2R3Q
 
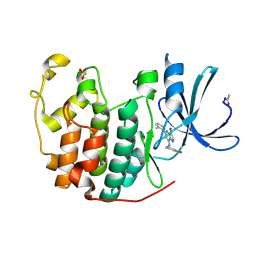 | | Crystal Structure of Cyclin-Dependent Kinase 2 with inhibitor | | Descriptor: | 3-((3-bromo-5-o-tolylpyrazolo[1,5-a]pyrimidin-7-ylamino)methyl)pyridine 1-oxide, Cell division protein kinase 2 | | Authors: | Fischmann, T.O, Hruza, A.W, Madison, V.M, Duca, J.S. | | Deposit date: | 2007-08-29 | | Release date: | 2008-01-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structure-guided discovery of cyclin-dependent kinase inhibitors.
Biopolymers, 89, 2008
|
|
2R3R
 
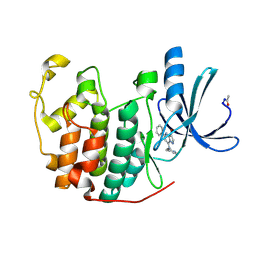 | | Crystal Structure of Cyclin-Dependent Kinase 2 with inhibitor | | Descriptor: | 3-bromo-5-phenyl-N-(pyridin-4-ylmethyl)pyrazolo[1,5-a]pyrimidin-7-amine, Cell division protein kinase 2 | | Authors: | Fischmann, T.O, Hruza, A.W, Madison, V.M, Duca, J.S. | | Deposit date: | 2007-08-29 | | Release date: | 2008-01-22 | | Last modified: | 2021-07-28 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Structure-guided discovery of cyclin-dependent kinase inhibitors.
Biopolymers, 89, 2008
|
|
2R3S
 
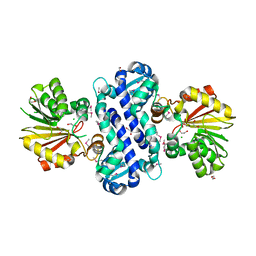 | |
2R3T
 
 | | I50V HIV-1 protease mutant in complex with a carbamoyl decorated pyrrolidine-based inhibitor | | Descriptor: | 4,4'-{(3S,4S)-PYRROLIDINE-3,4-DIYLBIS[(BENZYLIMINO)SULFONYL]}DIBENZAMIDE, CHLORIDE ION, Protease | | Authors: | Boettcher, J, Blum, A, Heine, A, Diederich, W.E, Klebe, G. | | Deposit date: | 2007-08-30 | | Release date: | 2008-09-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and Kinetic Analysis of Pyrrolidine-Based Inhibitors of the Drug-Resistant Ile84Val Mutant of HIV-1 Protease
J.Mol.Biol., 383, 2008
|
|
2R3U
 
 | | Crystal structure of the PDZ deletion mutant of DegS | | Descriptor: | Protease degS | | Authors: | Clausen, T, Kurzbauer, R. | | Deposit date: | 2007-08-30 | | Release date: | 2007-11-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Regulation of the sigmaE stress response by DegS: how the PDZ domain keeps the protease inactive in the resting state and allows integration of different OMP-derived stress signals upon folding stress.
Genes Dev., 21, 2007
|
|
2R3V
 
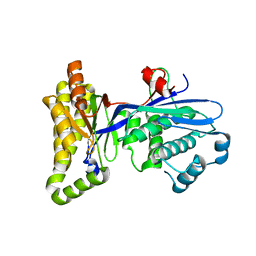 | |
2R3W
 
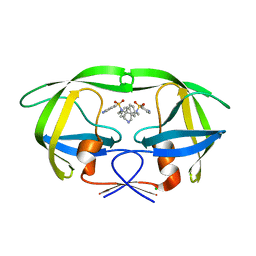 | | I84V HIV-1 protease in complex with a amino decorated pyrrolidine-based inhibitor | | Descriptor: | CHLORIDE ION, N,N'-(3S,4S)-PYRROLIDINE-3,4-DIYLBIS(4-AMINO-N-BENZYLBENZENESULFONAMIDE), Protease | | Authors: | Boettcher, J, Blum, A, Heine, A, Diederich, W.E, Klebe, G. | | Deposit date: | 2007-08-30 | | Release date: | 2008-09-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structural and Kinetic Analysis of Pyrrolidine-Based Inhibitors of the Drug-Resistant Ile84Val Mutant of HIV-1 Protease
J.Mol.Biol., 383, 2008
|
|
2R3X
 
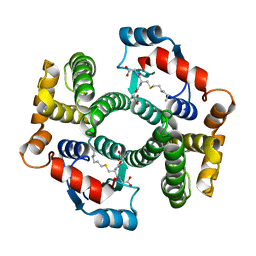 | | Crystal structure of an R15L hGSTA1-1 mutant complexed with S-hexyl-glutathione | | Descriptor: | Glutathione S-transferase A1, S-HEXYLGLUTATHIONE | | Authors: | Burke, J.P.W.G, Kinsley, N, Sayed, M, Sewell, T, Dirr, H.W. | | Deposit date: | 2007-08-30 | | Release date: | 2007-12-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Arginine 15 stabilizes an S(N)Ar reaction transition state and the binding of anionic ligands at the active site of human glutathione transferase A1-1.
Biophys.Chem., 146, 2010
|
|
2R3Y
 
 | |
2R3Z
 
 | | Crystal structure of mouse IP-10 | | Descriptor: | Small-inducible cytokine B10 | | Authors: | Jabeen, T, Leonard, P, Jamaluddin, H, Acharya, K.R. | | Deposit date: | 2007-08-30 | | Release date: | 2008-08-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of mouse IP-10, a chemokine
Acta Crystallogr.,Sect.D, 64, 2008
|
|
2R40
 
 | | Crystal structure of 20E bound EcR/USP | | Descriptor: | (2beta,3beta,5beta,22R)-2,3,14,20,22,25-hexahydroxycholest-7-en-6-one, CITRATE ANION, Ecdysone Receptor, ... | | Authors: | Moras, D, Billas, I.M.L, Browning, C. | | Deposit date: | 2007-08-30 | | Release date: | 2007-10-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.402 Å) | | Cite: | Critical Role of Desolvation in the Binding of 20-Hydroxyecdysone to the Ecdysone Receptor
J.Biol.Chem., 282, 2007
|
|
2R41
 
 | |
2R42
 
 | | The Biochemical and Structural Basis for feedback Inhibition of Mevalonate Kinase and Isoprenoid Metabolism | | Descriptor: | MAGNESIUM ION, Mevalonate kinase, S-[(2E,6E)-3,7,11-TRIMETHYLDODECA-2,6,10-TRIENYL] TRIHYDROGEN THIODIPHOSPHATE | | Authors: | Fu, Z, Voynova, N.E, Miziorko, H.M, Kim, J.P. | | Deposit date: | 2007-08-30 | | Release date: | 2008-06-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Biochemical and Structural Basis for Feedback Inhibition of Mevalonate Kinase and Isoprenoid Metabolism.
Biochemistry, 47, 2008
|
|
2R43
 
 | | I50V HIV-1 protease in complex with an amino decorated pyrrolidine-based inhibitor | | Descriptor: | CHLORIDE ION, GLYCEROL, N,N'-(3S,4S)-PYRROLIDINE-3,4-DIYLBIS(4-AMINO-N-BENZYLBENZENESULFONAMIDE), ... | | Authors: | Boettcher, J, Blum, A, Heine, A, Diederich, W.E, Klebe, G. | | Deposit date: | 2007-08-30 | | Release date: | 2008-09-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Structural and Kinetic Analysis of Pyrrolidine-Based Inhibitors of the Drug-Resistant Ile84Val Mutant of HIV-1 Protease
J.Mol.Biol., 383, 2008
|
|
2R44
 
 | |
