8DLM
 
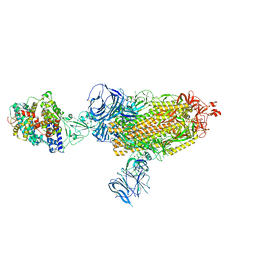 | | Cryo-EM structure of SARS-CoV-2 Beta (B.1.351) spike protein in complex with human ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Zhu, X, Mannar, D, Saville, J.W, Srivastava, S.S, Berezuk, A.M, Zhou, S, Tuttle, K.S, Subramaniam, S. | | Deposit date: | 2022-07-08 | | Release date: | 2022-08-31 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | SARS-CoV-2 variants of concern: spike protein mutational analysis and epitope for broad neutralization.
Nat Commun, 13, 2022
|
|
7FZ1
 
 | | Crystal Structure of human FABP4 in complex with 3-[(E)-anilino-(2-oxo-1H-indol-3-ylidene)methyl]sulfanylpropanoic acid | | Descriptor: | 3-{[(E)-anilino(2-oxo-1,2-dihydro-3H-indol-3-ylidene)methyl]sulfanyl}propanoic acid, FORMIC ACID, Fatty acid-binding protein, ... | | Authors: | Ehler, A, Benz, J, Obst, U, Mischke, S, Rudolph, M.G. | | Deposit date: | 2023-04-27 | | Release date: | 2023-06-14 | | Last modified: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (0.99 Å) | | Cite: | A high-resolution data set of fatty acid-binding protein structures. III. Unexpectedly high occurrence of wrong ligands.
Acta Crystallogr D Struct Biol, 81, 2025
|
|
5F6J
 
 | |
6RIV
 
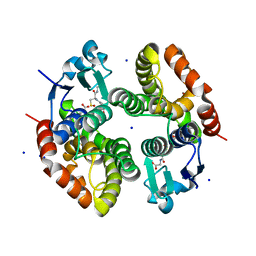 | | Crystal structure of Alopecurus myosuroides GSTF | | Descriptor: | GLYCEROL, Glutathione transferase, S-Hydroxy-Glutathione, ... | | Authors: | Papageorgiou, A.C, Poudel, N. | | Deposit date: | 2019-04-25 | | Release date: | 2020-03-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Comparative structural and functional analysis of phi class glutathione transferases involved in multiple-herbicide resistance of grass weeds and crops.
Plant Physiol Biochem., 149, 2020
|
|
7K88
 
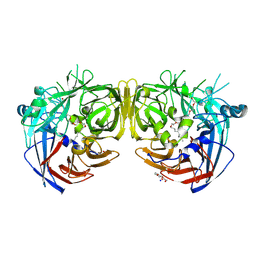 | | Crystal structure of bovine RPE65 in complex with hexaethylene glycol monooctyl ether | | Descriptor: | 3,6,9,12,15,18-hexaoxahexacosan-1-ol, FE (II) ION, Retinoid isomerohydrolase, ... | | Authors: | Kiser, P.D. | | Deposit date: | 2020-09-26 | | Release date: | 2021-06-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Rational Alteration of Pharmacokinetics of Chiral Fluorinated and Deuterated Derivatives of Emixustat for Retinal Therapy.
J.Med.Chem., 64, 2021
|
|
3H4C
 
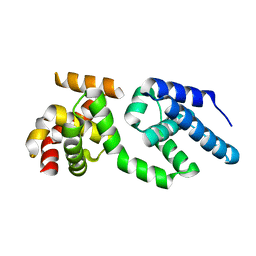 | | Structure of the C-terminal Domain of Transcription Factor IIB from Trypanosoma brucei | | Descriptor: | 1,2-ETHANEDIOL, Transcription factor TFIIB-like | | Authors: | Syed Ibrahim, B, Kanneganti, N, Rieckhof, G.E, Das, A, Laurents, D.V, Palenchar, J.B, Bellofatto, V, Wah, D.A. | | Deposit date: | 2009-04-18 | | Release date: | 2009-08-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the C-terminal domain of transcription factor IIB from Trypanosoma brucei.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
6ICR
 
 | | LdCoroCC mutant- C482A | | Descriptor: | 1,2-ETHANEDIOL, Coronin-like protein | | Authors: | Karade, S.S, Srivastava, V.K, Ansari, A, Pratap, J.V. | | Deposit date: | 2018-09-06 | | Release date: | 2019-10-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Molecular and structural analysis of a mechanical transition of helices in the L. donovani coronin coiled-coil domain.
Int.J.Biol.Macromol., 143, 2020
|
|
5BQN
 
 | | Crystal structure of the LHn fragment of botulinum neurotoxin type D, mutant H233Y E230Q | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Botulinum neurotoxin type D,Botulinum neurotoxin type D | | Authors: | Masuyer, G, Davies, J.R, Moore, K, Chaddock, J.A, Acharya, K.R. | | Deposit date: | 2015-05-29 | | Release date: | 2015-08-19 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural analysis of Clostridium botulinum neurotoxin type D as a platform for the development of targeted secretion inhibitors.
Sci Rep, 5, 2015
|
|
7SZA
 
 | |
9I7Z
 
 | | LecA in complex with 2-fluoro non-carbohydrate glycomimetic | | Descriptor: | 1,2-ETHANEDIOL, 2-[2-[2-[2-[2-[2-(2-methoxyethoxy)ethoxy]ethoxy]ethoxy]ethoxy]ethoxy]ethanol, ACETATE ION, ... | | Authors: | Varrot, A. | | Deposit date: | 2025-02-03 | | Release date: | 2025-10-22 | | Last modified: | 2025-11-12 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The Parkinson's Disease Drug Tolcapone and Analogues are Potent Glycomimetic Lectin Inhibitors of Pseudomonas aeruginosa LecA.
Angew.Chem.Int.Ed.Engl., 2025
|
|
4Q99
 
 | |
6P89
 
 | | E.coli LpxD in complex with compound 7 | | Descriptor: | 3-hydroxy-7,7-dimethyl-2-phenyl-4-(thiophen-2-yl)-2,6,7,8-tetrahydro-5H-pyrazolo[3,4-b]quinolin-5-one, MAGNESIUM ION, N-(2,3-dihydro-1,4-benzodioxin-6-yl)-2-(8-methyl[1]benzopyrano[4,3-c]pyrazol-1(4H)-yl)acetamide, ... | | Authors: | Ma, X, Shia, S. | | Deposit date: | 2019-06-06 | | Release date: | 2019-08-28 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural and Biological Basis of Small Molecule Inhibition ofEscherichia coliLpxD Acyltransferase Essential for Lipopolysaccharide Biosynthesis.
Acs Infect Dis., 6, 2020
|
|
5L9H
 
 | | Crystal structure of T. brucei PDE-B1 catalytic domain with inhibitor NPD-340 | | Descriptor: | 5-[4-methoxy-3-[4-[4-(1~{H}-1,2,3,4-tetrazol-5-yl)phenoxy]butoxy]phenyl]-4,4-dimethyl-2-propan-2-yl-pyrazol-3-one, FORMIC ACID, GLYCEROL, ... | | Authors: | Singh, A.K, Brown, D.G. | | Deposit date: | 2016-06-10 | | Release date: | 2018-03-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Targeting a Subpocket in Trypanosoma brucei Phosphodiesterase B1 (TbrPDEB1) Enables the Structure-Based Discovery of Selective Inhibitors with Trypanocidal Activity.
J. Med. Chem., 61, 2018
|
|
7ZXA
 
 | | Crystal structure of human cathepsin L with covalently bound aloxistatin (E-64D) | | Descriptor: | 1,2-ETHANEDIOL, Cathepsin L, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Falke, S, Lieske, J, Guenther, S, Reinke, P.Y.A, Ewert, W, Loboda, J, Karnicar, K, Usenik, A, Lindic, N, Sekirnik, A, Chapman, H.N, Hinrichs, W, Turk, D, Meents, A. | | Deposit date: | 2022-05-20 | | Release date: | 2023-05-31 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural Elucidation and Antiviral Activity of Covalent Cathepsin L Inhibitors.
J.Med.Chem., 67, 2024
|
|
8TP3
 
 | | H1 hemagglutinin (NC99) in complex with RBS-targeting Fab 1-1-1F05 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of Fab 1-1-1F05, ... | | Authors: | Yang, Y.R, Han, J, Perrett, H.R, Ward, A.B. | | Deposit date: | 2023-08-04 | | Release date: | 2023-10-11 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Immune memory shapes human polyclonal antibody responses to H2N2 vaccination.
Cell Rep, 43, 2024
|
|
8DLL
 
 | | Cryo-EM structure of SARS-CoV-2 Beta (B.1.351) spike protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhu, X, Mannar, D, Saville, J.W, Srivastava, S.S, Berezuk, A.M, Zhou, S, Tuttle, K.S, Subramaniam, S. | | Deposit date: | 2022-07-08 | | Release date: | 2022-08-31 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (2.56 Å) | | Cite: | SARS-CoV-2 variants of concern: spike protein mutational analysis and epitope for broad neutralization.
Nat Commun, 13, 2022
|
|
5FAL
 
 | | Crystal structure of PvHCT in complex with CoA and p-coumaroyl-shikimate | | Descriptor: | (3~{R},4~{R},5~{R})-5-[(~{E})-3-(4-hydroxyphenyl)prop-2-enoyl]oxy-3,4-bis(oxidanyl)cyclohexene-1-carboxylic acid, COENZYME A, GLYCEROL, ... | | Authors: | Pereira, J.H, Moriarty, N.W, Eudes, A, Yogiswara, S, Wang, G, Benites, V.T, Baidoo, E.E.K, Lee, T.S, Keasling, J.D, Loque, D, Adams, P.D. | | Deposit date: | 2015-12-11 | | Release date: | 2016-02-24 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.861 Å) | | Cite: | Exploiting the Substrate Promiscuity of Hydroxycinnamoyl-CoA:Shikimate Hydroxycinnamoyl Transferase to Reduce Lignin.
Plant Cell.Physiol., 57, 2016
|
|
1LSU
 
 | | KTN Bsu222 Crystal Structure in Complex with NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Conserved hypothetical protein yuaA | | Authors: | Roosild, T.P, Miller, S, Booth, I.R, Choe, S. | | Deposit date: | 2002-05-18 | | Release date: | 2002-07-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | A mechanism of regulating transmembrane potassium flux through a ligand-mediated conformational switch.
Cell(Cambridge,Mass.), 109, 2002
|
|
6IWG
 
 | | Crystal structure of rhesus macaque MHC class I molecule Mamu-B*05104 complexed with N-myristoylated 4-mer lipopeptide derived from SIV nef protein | | Descriptor: | 1,2-ETHANEDIOL, BORIC ACID, Beta-2-microglobulin, ... | | Authors: | Yamamoto, Y, Morita, D, Sugita, M. | | Deposit date: | 2018-12-05 | | Release date: | 2019-08-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Identification and Structure of an MHC Class I-Encoded Protein with the Potential to PresentN-Myristoylated 4-mer Peptides to T Cells.
J Immunol., 202, 2019
|
|
9FF3
 
 | | The structure of delta-ScoC, a global regulator protein from Geobacillus kaustophilus T-1 | | Descriptor: | HTH-type transcriptional regulator Hpr | | Authors: | Hadad, N, Shulami, S, Pomyalov, S, Shoham, Y, Shoham, G. | | Deposit date: | 2024-05-22 | | Release date: | 2025-06-04 | | Method: | X-RAY DIFFRACTION (3.32 Å) | | Cite: | The structure of delta-ScoC, a global regulator protein from Geobacillus kaustophilus T-1
To Be Published
|
|
5LAH
 
 | | NMR structure of the sea anemone peptide tau-AnmTx Ueq 12-1 with an uncommon fold | | Descriptor: | tau-AnmTx Ueq 12-1 | | Authors: | Mineev, K.S, Arseniev, A.S, Andreev, Y.A, Kozlov, S.A, Logashina, Y.A. | | Deposit date: | 2016-06-14 | | Release date: | 2017-05-10 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | New Disulfide-Stabilized Fold Provides Sea Anemone Peptide to Exhibit Both Antimicrobial and TRPA1 Potentiating Properties
Toxins, 9, 2017
|
|
7T4T
 
 | | Crystal Structure of cGMP-dependent Protein Kinase | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, ... | | Authors: | Zebisch, M, Silvestre, L, Fischmann, T.O. | | Deposit date: | 2021-12-10 | | Release date: | 2023-06-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Selective small molecule activation of PKG1alpha: structure and function
Commun Biol, 2023
|
|
8QWL
 
 | | Structure of p53 cancer mutant Y163C | | Descriptor: | 1,2-ETHANEDIOL, Cellular tumor antigen p53, MALONATE ION, ... | | Authors: | Balourdas, D.I, Markl, A.M, Kraemer, A, Knapp, S, Joerger, A.C, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-10-19 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural basis of p53 inactivation by cavity-creating cancer mutations and its implications for the development of mutant p53 reactivators.
Cell Death Dis, 15, 2024
|
|
5INC
 
 | | Crystal structure of HLA-B5801, a protective HLA allele for HIV-1 infection | | Descriptor: | Beta-2-microglobulin, GLN-ALA-THR-GLN-GLU-VAL-LYS-ASN-TRP, HLA class I histocompatibility antigen, ... | | Authors: | Li, X, Wang, J.-H. | | Deposit date: | 2016-03-07 | | Release date: | 2016-10-05 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.881 Å) | | Cite: | Crystal structure of HLA-B*5801, a protective HLA allele for HIV-1 infection.
Protein Cell, 7, 2016
|
|
8FJP
 
 | | Cryo-EM structure of native mosquito salivary gland surface protein 1 (SGS1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, salivary gland surface protein 1 | | Authors: | Liu, S, Xia, X, Calvo, E, Zhou, Z.H. | | Deposit date: | 2022-12-20 | | Release date: | 2023-03-01 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Native structure of mosquito salivary protein uncovers domains relevant to pathogen transmission.
Nat Commun, 14, 2023
|
|
