6TDA
 
 | | Structure of SWI/SNF chromatin remodeler RSC bound to a nucleosome | | Descriptor: | Actin-like protein ARP9, Actin-related protein 7, Chromatin structure-remodeling complex protein RSC58, ... | | Authors: | Wagner, F.R, Dienemann, C, Wang, H, Stuetzer, A, Tegunov, D, Urlaub, H, Cramer, P. | | Deposit date: | 2019-11-08 | | Release date: | 2020-03-18 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (15 Å) | | Cite: | Structure of SWI/SNF chromatin remodeller RSC bound to a nucleosome.
Nature, 579, 2020
|
|
1U6G
 
 | | Crystal Structure of The Cand1-Cul1-Roc1 Complex | | Descriptor: | Cullin homolog 1, RING-box protein 1, TIP120 protein, ... | | Authors: | Goldenberg, S.J, Shumway, S.D, Cascio, T.C, Garbutt, K.C, Liu, J, Xiong, Y, Zheng, N. | | Deposit date: | 2004-07-29 | | Release date: | 2004-12-14 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of the Cand1-Cul1-Roc1 complex reveals regulatory mechanisms for the assembly of the multisubunit cullin-dependent ubiquitin ligases
Cell(Cambridge,Mass.), 119, 2004
|
|
6KW5
 
 | | The ClassC RSC-Nucleosome Complex | | Descriptor: | Actin-like protein ARP9, Actin-related protein 7, Chromatin structure-remodeling complex protein RSC3, ... | | Authors: | Ye, Y.P, Wu, H, Chen, K.J, Verma, N, Cairns, B, Gao, N, Chen, Z.C. | | Deposit date: | 2019-09-06 | | Release date: | 2020-09-09 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (10.13 Å) | | Cite: | Structure of the RSC complex bound to the nucleosome
To Be Published
|
|
6KW4
 
 | | The ClassB RSC-Nucleosome Complex | | Descriptor: | Actin-like protein ARP9, Actin-related protein 7, Chromatin structure-remodeling complex protein RSC3, ... | | Authors: | Ye, Y.P, Wu, H, Chen, K.J, Verma, N, Cairns, B, Gao, N, Chen, Z.C. | | Deposit date: | 2019-09-06 | | Release date: | 2019-11-13 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (7.55 Å) | | Cite: | Structure of the RSC complex bound to the nucleosome.
Science, 366, 2019
|
|
7N4K
 
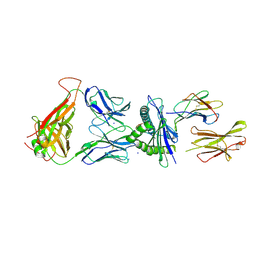 | | 6218 TCR in complex with H2-Db PA 224 | | Descriptor: | Beta-2-microglobulin, Fusion protein of T cell receptor alpha variable 21-DV12 and T-cell receptor, sp3.4 alpha chain, ... | | Authors: | Szeto, C, Gras, S. | | Deposit date: | 2021-06-04 | | Release date: | 2022-07-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Covalent TCR-peptide-MHC interactions induce T cell activation and redirect T cell fate in the thymus.
Nat Commun, 13, 2022
|
|
7N5P
 
 | | 6218 TCR in complex with H2-Db PA224-233 with a cysteine mutant | | Descriptor: | Beta-2-microglobulin, Fusion protein of T cell receptor alpha variable 21-DV12 and T-cell receptor, sp3.4 alpha chain, ... | | Authors: | Szeto, C, Gras, S. | | Deposit date: | 2021-06-06 | | Release date: | 2022-07-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Covalent TCR-peptide-MHC interactions induce T cell activation and redirect T cell fate in the thymus.
Nat Commun, 13, 2022
|
|
7N5Q
 
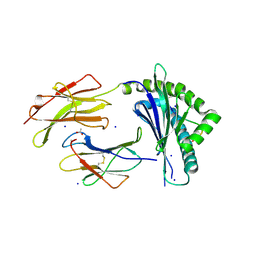 | |
5XM1
 
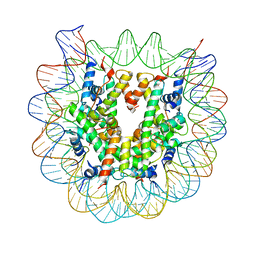 | | The mouse nucleosome structure containing H2A, H2B type3-A, H3mm7, and H4 | | Descriptor: | DNA (146-MER), Histone H2A type 1-B, Histone H2B type 3-A, ... | | Authors: | Taguchi, H, Horikoshi, N, Kurumizaka, H. | | Deposit date: | 2017-05-12 | | Release date: | 2018-03-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | Histone H3.3 sub-variant H3mm7 is required for normal skeletal muscle regeneration.
Nat Commun, 9, 2018
|
|
8HY0
 
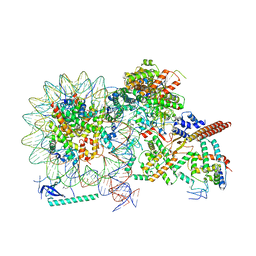 | |
8HXY
 
 | |
3K3J
 
 | | P38alpha bound to novel DFG-out compound PF-00416121 | | Descriptor: | 2-(4-fluorophenyl)-3-oxo-6-pyridin-4-yl-N-[2-(trifluoromethyl)benzyl]-2,3-dihydropyridazine-4-carboxamide, 2-fluoro-4-[4-(4-fluorophenyl)-1H-pyrazol-3-yl]pyridine, Mitogen-activated protein kinase 14 | | Authors: | Kazmirski, S.L, DiNitto, J.P. | | Deposit date: | 2009-10-02 | | Release date: | 2009-11-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.995 Å) | | Cite: | The Design, Synthesis and Potential Utility of Fluorescence Probes that Target DFG-out Conformation of p38alpha for High Throughput Screening Binding Assay.
Chem.Biol.Drug Des., 74, 2009
|
|
4D18
 
 | | Crystal structure of the COP9 signalosome | | Descriptor: | COP9 SIGNALOSOME COMPLEX SUBUNIT 1, COP9 SIGNALOSOME COMPLEX SUBUNIT 2, COP9 SIGNALOSOME COMPLEX SUBUNIT 3, ... | | Authors: | Bunker, R.D, Lingaraju, G.M, Thoma, N.H. | | Deposit date: | 2014-05-01 | | Release date: | 2014-07-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (4.08 Å) | | Cite: | Crystal Structure of the Human Cop9 Signalosome
Nature, 512, 2014
|
|
3S3I
 
 | | p38 kinase crystal structure in complex with small molecule inhibitor | | Descriptor: | 3-(3-tert-butyl[1,2,4]triazolo[4,3-a]pyridin-7-yl)-N-cyclopropyl-4-methylbenzamide, Mitogen-activated protein kinase 14 | | Authors: | Segarra, V, Aiguade, J, Roca, R, Fisher, M, Lamers, M. | | Deposit date: | 2011-05-18 | | Release date: | 2012-04-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Novel triazolopyridylbenzamides as potent and selective p38 alpha inhibitors.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
3D7Z
 
 | |
1KV1
 
 | | p38 MAP Kinase in Complex with Inhibitor 1 | | Descriptor: | 1-(5-TERT-BUTYL-2-METHYL-2H-PYRAZOL-3-YL)-3-(4-CHLORO-PHENYL)-UREA, p38 MAP kinase | | Authors: | Pargellis, C, Tong, L, Churchill, L, Cirillo, P.F, Gilmore, T, Graham, A.G, Grob, P.M, Hickey, E.R, Moss, N, Pav, S, Regan, J. | | Deposit date: | 2002-01-23 | | Release date: | 2002-03-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Inhibition of p38 MAP kinase by utilizing a novel allosteric binding site.
Nat.Struct.Biol., 9, 2002
|
|
9MHB
 
 | | Crystal Structure of Human P38 alpha MAPK In Complex with MW01-14-064SRM | | Descriptor: | (5P)-N,N-dimethyl-5-(naphthalen-1-yl)-6-(pyridin-4-yl)pyrazin-2-amine, 1,2-ETHANEDIOL, 4-[3-(4-FLUOROPHENYL)-1H-PYRAZOL-4-YL]PYRIDINE, ... | | Authors: | Minasov, G, Tokars, V.L, Roy, S.M, Watterson, D.M, Shuvalova, L. | | Deposit date: | 2024-12-11 | | Release date: | 2025-01-01 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal Structure of Human P38 alpha MAPK In Complex with MW01-14-064SRM
To Be Published
|
|
1KV2
 
 | | Human p38 MAP Kinase in Complex with BIRB 796 | | Descriptor: | 1-(5-TERT-BUTYL-2-P-TOLYL-2H-PYRAZOL-3-YL)-3-[4-(2-MORPHOLIN-4-YL-ETHOXY)-NAPHTHALEN-1-YL]-UREA, p38 MAP kinase | | Authors: | Pargellis, C, Tong, L, Churchill, L, Cirillo, P.F, Gilmore, T, Graham, A.G, Grob, P.M, Hickey, E.R, Moss, N, Pav, S, Regan, J. | | Deposit date: | 2002-01-23 | | Release date: | 2002-03-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Inhibition of p38 MAP kinase by utilizing a novel allosteric binding site.
Nat.Struct.Biol., 9, 2002
|
|
2I0H
 
 | | The structure of p38alpha in complex with an arylpyridazinone | | Descriptor: | 2-(3-{(2-CHLORO-4-FLUOROPHENYL)[1-(2-CHLOROPHENYL)-6-OXO-1,6-DIHYDROPYRIDAZIN-3-YL]AMINO}PROPYL)-1H-ISOINDOLE-1,3(2H)-DIONE, GLYCEROL, Mitogen-activated protein kinase 14 | | Authors: | Natarajan, S.R, Heller, S.T, Nam, K, Singh, S.B, Scapin, G, Patel, S, Thompson, J.E, Fitzgerald, C.E, O'Keefe, S.J. | | Deposit date: | 2006-08-10 | | Release date: | 2006-10-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | p38 MAP Kinase Inhibitors Part 6: 2-Arylpyridazin-3-ones as templates for inhibitor design.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
5Y3R
 
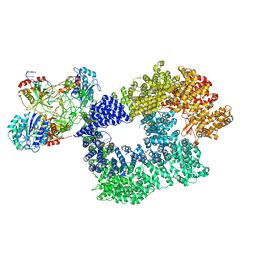 | | Cryo-EM structure of Human DNA-PK Holoenzyme | | Descriptor: | DNA (34-MER), DNA (36-MER), DNA-dependent protein kinase catalytic subunit, ... | | Authors: | Yin, X, Liu, M, Tian, Y, Wang, J, Xu, Y. | | Deposit date: | 2017-07-29 | | Release date: | 2017-09-06 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (6.6 Å) | | Cite: | Cryo-EM structure of human DNA-PK holoenzyme
Cell Res., 27, 2017
|
|
9LKY
 
 | | Dimer of CRL2-FEM1B bound with PLD6 | | Descriptor: | Cullin-2, E3 ubiquitin-protein ligase RBX1, N-terminally processed, ... | | Authors: | Zhao, S, Xu, C. | | Deposit date: | 2025-01-17 | | Release date: | 2025-04-09 | | Last modified: | 2025-05-07 | | Method: | ELECTRON MICROSCOPY (3.93 Å) | | Cite: | TOM20-driven E3 ligase recruitment regulates mitochondrial dynamics through PLD6.
Nat.Chem.Biol., 2025
|
|
8HXX
 
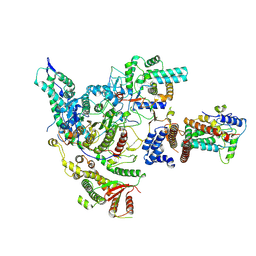 | | Cryo-EM structure of the histone deacetylase complex Rpd3S | | Descriptor: | Chromatin modification-related protein EAF3, Histone H3, Histone deacetylase RPD3, ... | | Authors: | Cui, H, Wang, H. | | Deposit date: | 2023-01-05 | | Release date: | 2023-09-27 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure of histone deacetylase complex Rpd3S bound to nucleosome.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8HXZ
 
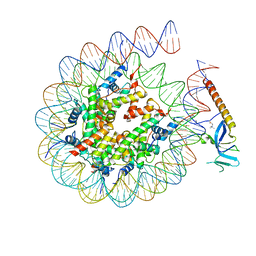 | | Cryo-EM structure of Eaf3 CHD in complex with nucleosome | | Descriptor: | Chromatin modification-related protein EAF3, DNA (352-MER), Histone H2A, ... | | Authors: | Cui, H, Wang, H. | | Deposit date: | 2023-01-05 | | Release date: | 2023-09-27 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure of histone deacetylase complex Rpd3S bound to nucleosome.
Nat.Struct.Mol.Biol., 30, 2023
|
|
9N6B
 
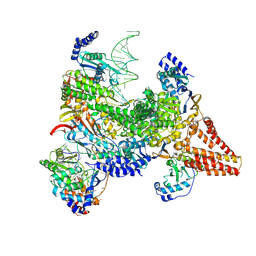 | | Structure of the retron IA complex with HNH nuclease in the "up" orientation | | Descriptor: | AAA family ATPase, ADENOSINE-5'-TRIPHOSPHATE, RNA-directed DNA polymerase, ... | | Authors: | Burman, N, Thomas-George, J, Wilkinson, R, Wiedenheft, B. | | Deposit date: | 2025-02-05 | | Release date: | 2025-04-23 | | Method: | ELECTRON MICROSCOPY (3.09 Å) | | Cite: | Structural basis of antiphage defense by an ATPase-associated reverse transcriptase.
Biorxiv, 2025
|
|
5XXD
 
 | |
5XXJ
 
 | |
