1PR2
 
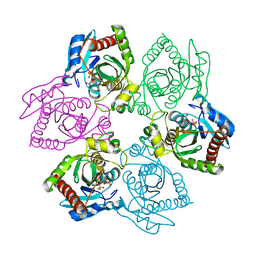 | | Escherichia coli Purine Nucleoside Phosphorylase Complexed with 9-beta-D-[2-deoxyribofuranosyl]-6-methylpurine and Phosphate/Sulfate | | Descriptor: | 9-(2-DEOXY-BETA-D-RIBOFURANOSYL)-6-METHYLPURINE, PHOSPHATE ION, Purine nucleoside phosphorylase DeoD-type | | Authors: | Bennett, E.M, Li, C, Allan, P.W, Parker, W.B, Ealick, S.E. | | Deposit date: | 2003-06-19 | | Release date: | 2003-11-25 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for substrate specificity of Escherichia coli purine nucleoside phosphorylase.
J.Biol.Chem., 278, 2003
|
|
1PTW
 
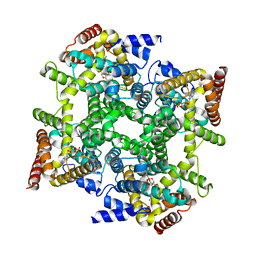 | |
1Q3W
 
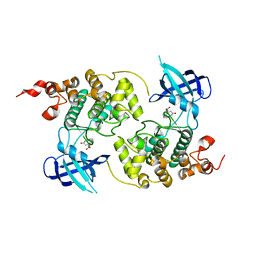 | | GSK-3 Beta complexed with Alsterpaullone | | Descriptor: | 9-NITRO-5,12-DIHYDRO-7H-BENZO[2,3]AZEPINO[4,5-B]INDOL-6-ONE, GLYCOGEN SYNTHASE KINASE-3 BETA | | Authors: | Bertrand, J.A, Thieffine, S, Vulpetti, A, Cristiani, C, Valsasina, B, Knapp, S, Kalisz, H.M, Flocco, M. | | Deposit date: | 2003-08-01 | | Release date: | 2003-10-21 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Characterization of the Gsk-3Beta Active Site Using Selective and Non-selective ATP-Mimetic Inhibitors
J.Mol.Biol., 333, 2003
|
|
1Q6S
 
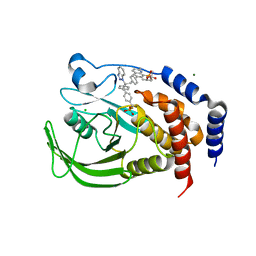 | | THE STRUCTURE OF PHOSPHOTYROSINE PHOSPHATASE 1B IN COMPLEX WITH COMPOUND 9 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 6-[4-((2R)-2-(1H-1,2,3-BENZOTRIAZOL-1-YL)-3-{4-[DIFLUORO(PHOSPHONO)METHYL]PHENYL}-2-PHENYLPROPYL)PHENYL]-2-METHYLQUINOLIN-8-YLPHOSPHONIC ACID, CHLORIDE ION, ... | | Authors: | Scapin, G, Patel, S.B, Becker, J.W, Wang, Q, Desponts, C, Waddleton, D, Skorey, K, Cromlish, W, Bayly, C, Therien, M, Gauthier, J.Y, Li, C.S, Lau, C.K, Ramachandran, C, Kennedy, B.P, Asante-Appiah, E. | | Deposit date: | 2003-08-13 | | Release date: | 2003-09-30 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Structural Basis for the Selectivity of Benzotriazole Inhibitors of Ptp1B
Biochemistry, 42, 2003
|
|
1Q8T
 
 | | The Catalytic Subunit of cAMP-dependent Protein Kinase (PKA) in Complex with Rho-kinase Inhibitor Y-27632 | | Descriptor: | (R)-TRANS-4-(1-AMINOETHYL)-N-(4-PYRIDYL) CYCLOHEXANECARBOXAMIDE, cAMP-dependent protein kinase inhibitor, alpha form, ... | | Authors: | Breitenlechner, C, Gassel, M, Hidaka, H, Kinzel, V, Huber, R, Engh, R.A, Bossemeyer, D. | | Deposit date: | 2003-08-22 | | Release date: | 2003-12-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Protein kinase A in complex with Rho-kinase inhibitors Y-27632, Fasudil, and H-1152P: structural basis of selectivity.
Structure, 11, 2003
|
|
1QYD
 
 | | Crystal structures of pinoresinol-lariciresinol and phenylcoumaran benzylic ether reductases, and their relationship to isoflavone reductases | | Descriptor: | pinoresinol-lariciresinol reductase | | Authors: | Min, T, Kasahara, H, Bedgar, D.L, Youn, B, Lawrence, P.K, Gang, D.R, Halls, S.C, Park, H, Hilsenbeck, J.L, Davin, L.B, Kang, C. | | Deposit date: | 2003-09-10 | | Release date: | 2003-11-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of pinoresinol-lariciresinol and phenylcoumaran benzylic ether reductases and their relationship to isoflavone reductases.
J.Biol.Chem., 278, 2003
|
|
1QNX
 
 | | Ves v 5, an allergen from Vespula vulgaris venom | | Descriptor: | SODIUM ION, VES V 5 | | Authors: | Henriksen, A, Gajhede, M, Spangfort, M.D. | | Deposit date: | 1999-10-25 | | Release date: | 2000-10-26 | | Last modified: | 2018-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Major Venom Allergen of Yellow Jackets, Ves V 5: Structural Characterization of a Pathogenesis-Related Protein Superfamily.
Proteins: Struct.,Funct., Genet., 45, 2001
|
|
1OAL
 
 | | Active site copper and zinc ions modulate the quaternary structure of prokaryotic Cu,Zn superoxide dismutase | | Descriptor: | COPPER (II) ION, SUPEROXIDE DISMUTASE, ZINC ION | | Authors: | Cioni, P, Pesce, A, Rocca, B.M.D, Castellifalconiparrilli, L, Bolognesi, M, Strambini, G, Desideri, A. | | Deposit date: | 2003-01-15 | | Release date: | 2003-02-27 | | Last modified: | 2019-10-09 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Active-Site Copper and Zinc Ions Modulate the Quaternary Structure of Prokaryotic Cu,Zn Superoxide Dismutase
J.Mol.Biol., 326, 2003
|
|
1PR4
 
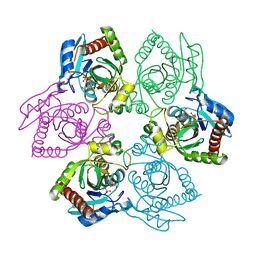 | | Escherichia coli Purine Nucleoside Phosphorylase Complexed with 9-beta-D-ribofuranosyl-6-methylthiopurine and Phosphate/Sulfate | | Descriptor: | 2-HYDROXYMETHYL-5-(6-METHYLSULFANYL-PURIN-9-YL)-TETRAHYDRO-FURAN-3,4-DIOL, PHOSPHATE ION, Purine nucleoside phosphorylase DeoD-type | | Authors: | Bennett, E.M, Li, C, Allan, P.W, Parker, W.B, Ealick, S.E. | | Deposit date: | 2003-06-19 | | Release date: | 2003-11-25 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for substrate specificity of Escherichia coli purine nucleoside phosphorylase.
J.Biol.Chem., 278, 2003
|
|
1PPO
 
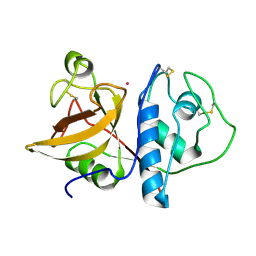 | | DETERMINATION OF THE STRUCTURE OF PAPAYA PROTEASE OMEGA | | Descriptor: | MERCURY (II) ION, PROTEASE OMEGA | | Authors: | Pickersgill, R.W, Rizkallah, P.J, Harris, G.W, Goodenough, P.W. | | Deposit date: | 1991-07-12 | | Release date: | 1993-10-31 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Determination of the Structure of Papaya Protease Omega
Acta Crystallogr.,Sect.B, 47, 1991
|
|
1PUO
 
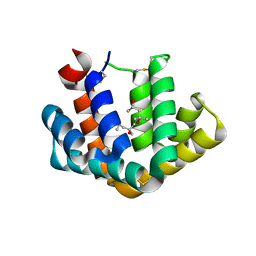 | | Crystal structure of Fel d 1- the major cat allergen | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Major allergen I polypeptide, fused chain 2, ... | | Authors: | Kaiser, L, Gronlund, H, Sandalova, T, Ljunggren, H.G, van Hage-Hamsten, M, Achour, A, Schneider, G. | | Deposit date: | 2003-06-25 | | Release date: | 2003-10-14 | | Last modified: | 2019-07-03 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The crystal structure of the major cat allergen Fel d 1, a member of the secretoglobin family.
J.Biol.Chem., 278, 2003
|
|
1PW7
 
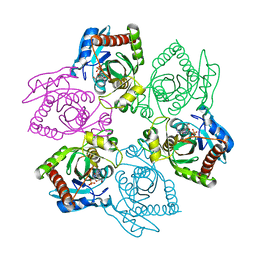 | | Crystal Structure of E. coli purine nucleoside phosphorylase complexed with 9-beta-D-arabinofuranosyladenine and sulfate/phosphate | | Descriptor: | 2-(6-AMINO-PURIN-9-YL)-5-HYDROXYMETHYL-TETRAHYDRO-FURAN-3,4-DIOL, PHOSPHATE ION, Purine nucleoside phosphorylase DeoD-type | | Authors: | Bennett, E.M, Li, C, Allan, P.W, Parker, W.B, Ealick, S.E. | | Deposit date: | 2003-06-30 | | Release date: | 2003-11-25 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for substrate specificity of Escherichia coli purine nucleoside phosphorylase.
J.Biol.Chem., 278, 2003
|
|
1Q4J
 
 | |
1Q4X
 
 | | Crystal Structure of Human Thyroid Hormone Receptor beta LBD in complex with specific agonist GC-24 | | Descriptor: | Thyroid hormone receptor beta-1, [4-(3-BENZYL-4-HYDROXYBENZYL)-3,5-DIMETHYLPHENOXY]ACETIC ACID | | Authors: | Borngraeber, S, Budny, M.J, Chiellini, G, Cunha-Lima, S.T, Togashi, M, Webb, P, Baxter, J.D, Scanlan, T.S, Fletterick, R.J. | | Deposit date: | 2003-08-04 | | Release date: | 2004-02-03 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Ligand selectivity by seeking hydrophobicity in thyroid hormone receptor.
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1PXH
 
 | | Crystal structure of protein tyrosine phosphatase 1B with potent and selective bidentate inhibitor compound 2 | | Descriptor: | ACETIC ACID, MAGNESIUM ION, N-{1-[5-(1-CARBAMOYL-2-MERCAPTO-ETHYLCARBAMOYL)-PENTYLCARBAMOYL]-2-[4-(DIFLUORO-PHOSPHONO-METHYL)-PHENYL]-ETHYL}-3-{2-[4-(DIFLUORO-PHOSPHONO-METHYL)-PHENYL]-ACETYLAMINO}-SUCCINAMIC ACID, ... | | Authors: | Sun, J.P, Fedorov, A, Lee, S.Y, Guo, X.L, Shen, K, Lawrence, D.S, Almo, S.C, Zhang, Z.Y. | | Deposit date: | 2003-07-04 | | Release date: | 2003-08-12 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of PTP1B complexed with a potent and selective bidentate inhibitor.
J.Biol.Chem., 278, 2003
|
|
1PR1
 
 | | Escherichia coli Purine Nucleoside Phosphorylase Complexed with Formycin B and Phosphate/Sulfate | | Descriptor: | FORMYCIN B, PHOSPHATE ION, Purine nucleoside phosphorylase | | Authors: | Bennett, E.M, Li, C, Allan, P.W, Parker, W.B, Ealick, S.E. | | Deposit date: | 2003-06-19 | | Release date: | 2003-11-25 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for substrate specificity of Escherichia coli purine nucleoside phosphorylase.
J.Biol.Chem., 278, 2003
|
|
1Q4L
 
 | | GSK-3 Beta complexed with Inhibitor I-5 | | Descriptor: | 2-CHLORO-5-[4-(3-CHLORO-PHENYL)-2,5-DIOXO-2,5-DIHYDRO-1H-PYRROL-3-YLAMINO]-BENZOIC ACID, GLYCOGEN SYNTHASE KINASE-3 BETA | | Authors: | Bertrand, J.A, Thieffine, S, Vulpetti, A, Cristiani, C, Valsasina, B, Knapp, S, Kalisz, H.M, Flocco, M. | | Deposit date: | 2003-08-04 | | Release date: | 2003-10-14 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Structural Characterization of the Gsk-3Beta Active Site Using Selective and Non-selective ATP-Mimetic Inhibitors
J.Mol.Biol., 333, 2003
|
|
1QX8
 
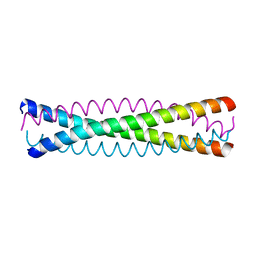 | | Crystal structure of a five-residue deletion mutant of the Rop protein | | Descriptor: | Regulatory protein ROP | | Authors: | Glykos, N.M, Vlassi, M, Papanikolaou, Y, Kotsifaki, D, Cesareni, G, Kokkinidis, M. | | Deposit date: | 2003-09-04 | | Release date: | 2004-09-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Loopless Rop: structure and dynamics of an engineered homotetrameric variant of the repressor of primer protein.
Biochemistry, 45, 2006
|
|
1R0A
 
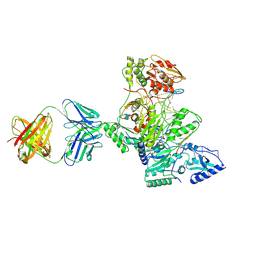 | | Crystal structure of HIV-1 reverse transcriptase covalently tethered to DNA template-primer solved to 2.8 angstroms | | Descriptor: | 5'-D(*A*TP*GP*CP*AP*TP*CP*GP*GP*CP*GP*CP*TP*CP*GP*AP*AP*CP*AP*GP*GP*GP*AP*CP*GP*GP*T)-3', 5'-D(*C*CP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*AP*GP*CP*GP*CP*CP*GP*(2DA))-3', GLYCEROL, ... | | Authors: | Tuske, S, Ding, J, Arnold, E. | | Deposit date: | 2003-09-19 | | Release date: | 2004-08-03 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Nonnucleoside inhibitor binding affects the interactions of the fingers subdomain of human immunodeficiency virus type 1 reverse transcriptase with DNA.
J.Virol., 78, 2004
|
|
1HVR
 
 | | RATIONAL DESIGN OF POTENT, BIOAVAILABLE, NONPEPTIDE CYCLIC UREAS AS HIV PROTEASE INHIBITORS | | Descriptor: | HIV-1 PROTEASE, [4R-(4ALPHA,5ALPHA,6BETA,7BETA)]-HEXAHYDRO-5,6-DIHYDROXY-1,3-BIS[2-NAPHTHYL-METHYL]-4,7-BIS(PHENYLMETHYL)-2H-1,3-DIAZEPIN-2-ONE | | Authors: | Chang, C.-H. | | Deposit date: | 1994-02-14 | | Release date: | 1995-01-26 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Rational design of potent, bioavailable, nonpeptide cyclic ureas as HIV protease inhibitors.
Science, 263, 1994
|
|
1EHW
 
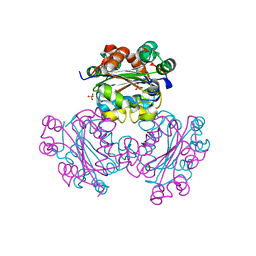 | | HUMAN NUCLEOSIDE DIPHOSPHATE KINASE 4 | | Descriptor: | NUCLEOSIDE DIPHOSPHATE KINASE, SULFATE ION | | Authors: | Milon, L, Meyer, P, Chiadmi, M, Munier, A, Johansson, M, Karlsson, A, Lascu, I, Capeau, J, Janin, J, Lacombe, M.-L. | | Deposit date: | 2000-02-23 | | Release date: | 2000-05-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The human nm23-H4 gene product is a mitochondrial nucleoside diphosphate kinase.
J.Biol.Chem., 275, 2000
|
|
1HW8
 
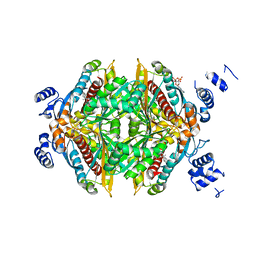 | | COMPLEX OF THE CATALYTIC PORTION OF HUMAN HMG-COA REDUCTASE WITH COMPACTIN (ALSO KNOWN AS MEVASTATIN) | | Descriptor: | (3R,5R)-3,5-dihydroxy-7-[(1S,2S,8S,8aR)-2-methyl-8-{[(2S)-2-methylbutanoyl]oxy}-1,2,6,7,8,8a-hexahydronaphthalen-1-yl]h eptanoic acid, ADENOSINE-5'-DIPHOSPHATE, HMG-COA REDUCTASE | | Authors: | Istvan, E.S, Deisenhofer, J. | | Deposit date: | 2001-01-09 | | Release date: | 2001-05-11 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural mechanism for statin inhibition of HMG-CoA reductase.
Science, 292, 2001
|
|
1EXX
 
 | | ENANTIOMER DISCRIMINATION ILLUSTRATED BY CRYSTAL STRUCTURES OF THE HUMAN RETINOIC ACID RECEPTOR HRARGAMMA LIGAND BINDING DOMAIN: THE COMPLEX WITH THE INACTIVE S-ENANTIOMER BMS270395. | | Descriptor: | 3-FLUORO-4-[2-HYDROXY-2-(5,5,8,8-TETRAMETHYL-5,6,7,8,-TETRAHYDRO-NAPHTALEN-2-YL)-ACETYLAMINO]-BENZOIC ACID, DODECYL-ALPHA-D-MALTOSIDE, RETINOIC ACID RECEPTOR GAMMA-2 | | Authors: | Klaholz, B.P, Mitschler, A, Belema, M, Zusi, C, Moras, D, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2000-05-05 | | Release date: | 2000-06-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Enantiomer discrimination illustrated by high-resolution crystal structures of the human nuclear receptor hRARgamma.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1HWL
 
 | |
1EPQ
 
 | | ENDOTHIA ASPARTIC PROTEINASE (ENDOTHIAPEPSIN) COMPLEXED WITH PD-133,450 (SOT PHE GLY+SCC GCL) | | Descriptor: | ENDOTHIAPEPSIN, N-[(1S)-2-{[(2S,3R,4S)-1-cyclohexyl-3,4-dihydroxy-6-methylheptan-2-yl]amino}-1-(ethylsulfanyl)-2-oxoethyl]-Nalpha-(morpholin-4-ylsulfonyl)-L-phenylalaninamide, SULFATE ION | | Authors: | Dealwis, C, Cooper, J.B, Blundell, T.L. | | Deposit date: | 1994-07-27 | | Release date: | 1994-12-20 | | Last modified: | 2020-05-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Analyses of ligand binding in five endothiapepsin crystal complexes and their use in the design and evaluation of novel renin inhibitors.
J.Med.Chem., 36, 1993
|
|
