8F6M
 
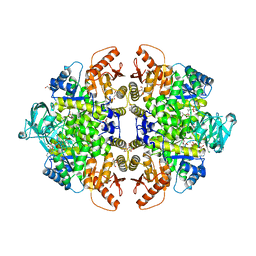 | | Complex of Rabbit muscle pyruvate kinase with ADP and the phosphonate analogue of PEP mimicking the Michaelis complex. | | Descriptor: | (E)-2-METHYL-3-PHOSPHONOACRYLATE, ADENOSINE-5'-DIPHOSPHATE, ALANINE, ... | | Authors: | Holyoak, T, Fenton, A.W. | | Deposit date: | 2022-11-16 | | Release date: | 2023-05-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | PYK-SubstitutionOME: an integrated database containing allosteric coupling, ligand affinity and mutational, structural, pathological, bioinformatic and computational information about pyruvate kinase isozymes.
Database (Oxford), 2023, 2023
|
|
8F6B
 
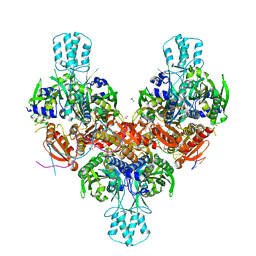 | | Crystal structure of murine PolG2 hexamer bound to DNA | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, DNA (5'-D(*CP*TP*GP*GP*TP*AP*GP*GP*CP*GP*CP*CP*TP*AP*CP*CP*AP*G)-3'), ... | | Authors: | Wojtaszek, J.L, Hoff, K.E, Williams, R.S. | | Deposit date: | 2022-11-16 | | Release date: | 2023-08-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structure-specific roles for PolG2-DNA complexes in maintenance and replication of mitochondrial DNA.
Nucleic Acids Res., 51, 2023
|
|
8F6A
 
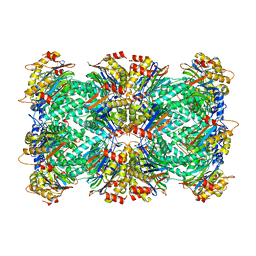 | | Thermoplasma acidophilum 20S proteasome - wild type | | Descriptor: | Proteasome subunit alpha, Proteasome subunit beta | | Authors: | Chuah, J, Smith, D. | | Deposit date: | 2022-11-16 | | Release date: | 2023-08-30 | | Method: | ELECTRON MICROSCOPY (2.06 Å) | | Cite: | High resolution structures define divergent and convergent mechanisms of archaeal proteasome activation.
Commun Biol, 6, 2023
|
|
8F66
 
 | |
8F61
 
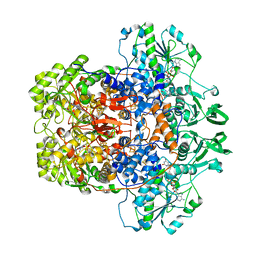 | | Dihydropyrimidine Dehydrogenase (DPD) C671S Mutant Soaked with Dihydrothymine Quasi-Anaerobically | | Descriptor: | (5S)-5-methyl-1,3-diazinane-2,4-dione, 1-DEOXY-1-(7,8-DIMETHYL-2,4-DIOXO-3,4-DIHYDRO-2H-BENZO[G]PTERIDIN-1-ID-10(5H)-YL)-5-O-PHOSPHONATO-D-RIBITOL, Dihydropyrimidine dehydrogenase [NADP(+)], ... | | Authors: | Kaley, N, Smith, M, Forouzesh, D, Liu, D, Moran, G. | | Deposit date: | 2022-11-15 | | Release date: | 2023-02-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Mammalian dihydropyrimidine dehydrogenase: Added mechanistic details from transient-state analysis of charge transfer complexes.
Arch.Biochem.Biophys., 736, 2023
|
|
8F5Z
 
 | | Composite map of CryoEM structure of Arabidopsis thaliana phytochrome A | | Descriptor: | 3-[5-[[(3~{R},4~{R})-3-ethyl-4-methyl-5-oxidanylidene-3,4-dihydropyrrol-2-yl]methyl]-2-[[5-[(4-ethyl-3-methyl-5-oxidanylidene-pyrrol-2-yl)methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-1~{H}-pyrrol-2-yl]methyl]-4-methyl-1~{H}-pyrrol-3-yl]propanoic acid, Phytochrome A | | Authors: | Li, H, Li, H. | | Deposit date: | 2022-11-15 | | Release date: | 2023-06-28 | | Last modified: | 2023-08-02 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | The structure of Arabidopsis phytochrome A reveals topological and functional diversification among the plant photoreceptor isoforms.
Nat.Plants, 9, 2023
|
|
8F5Y
 
 | | Crystal structure of pregnane X receptor ligand binding domain complexed with JQ1 | | Descriptor: | (6S)-6-(2-tert-butoxy-2-oxoethyl)-4-(4-chlorophenyl)-2,3,9-trimethyl-6,7-dihydrothieno[3,2-f][1,2,4]triazolo[4,3-a][1,4]diazepin-10-ium, Nuclear receptor coactivator 1, Nuclear receptor subfamily 1 group I member 2 | | Authors: | Huber, A.D, Poudel, S, Seetharaman, J, Miller, D.J, Chen, T. | | Deposit date: | 2022-11-15 | | Release date: | 2024-02-21 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | A bromodomain-independent mechanism of gene regulation by the BET inhibitor JQ1: direct activation of nuclear receptor PXR.
Nucleic Acids Res., 52, 2024
|
|
8F5X
 
 | | Crystal structure of human eosinophil-derived neurotoxin (EDN, ribonuclease 2) in complex with 5'-adenosine monophosphate (AMP) | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE MONOPHOSPHATE, Non-secretory ribonuclease, ... | | Authors: | Tran, T.T.Q, Pham, N.T.H, Calmettes, C, Doucet, N. | | Deposit date: | 2022-11-15 | | Release date: | 2023-11-29 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Ancestral sequence reconstruction dissects structural and functional differences among eosinophil ribonucleases.
J.Biol.Chem., 300, 2024
|
|
8F5W
 
 | | Dihydropyrimidine Dehydrogenase (DPD) C671S Mutant Soaked with Dihydrothymine and NADPH Quasi-Anaerobically | | Descriptor: | 1-DEOXY-1-(7,8-DIMETHYL-2,4-DIOXO-3,4-DIHYDRO-2H-BENZO[G]PTERIDIN-1-ID-10(5H)-YL)-5-O-PHOSPHONATO-D-RIBITOL, Dihydropyrimidine dehydrogenase [NADP(+)], FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Kaley, N, Smith, M, Forouzesh, D, Liu, D, Moran, G. | | Deposit date: | 2022-11-15 | | Release date: | 2023-02-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Mammalian dihydropyrimidine dehydrogenase: Added mechanistic details from transient-state analysis of charge transfer complexes.
Arch.Biochem.Biophys., 736, 2023
|
|
8F5U
 
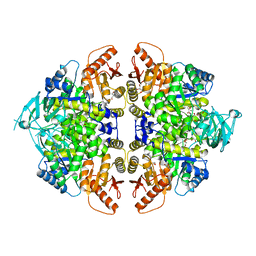 | | Rabbit muscle pyruvate kinase in complex with magnesium, potassium and pyruvate | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, ... | | Authors: | Holyoak, T, Fenton, A.W. | | Deposit date: | 2022-11-15 | | Release date: | 2023-05-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | PYK-SubstitutionOME: an integrated database containing allosteric coupling, ligand affinity and mutational, structural, pathological, bioinformatic and computational information about pyruvate kinase isozymes.
Database (Oxford), 2023, 2023
|
|
8F5T
 
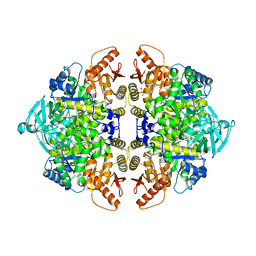 | | Rabbit muscle pyruvate kinase in complex with sodium and magnesium | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Holyoak, T, Fenton, A.W. | | Deposit date: | 2022-11-15 | | Release date: | 2023-05-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | PYK-SubstitutionOME: an integrated database containing allosteric coupling, ligand affinity and mutational, structural, pathological, bioinformatic and computational information about pyruvate kinase isozymes.
Database (Oxford), 2023, 2023
|
|
8F5N
 
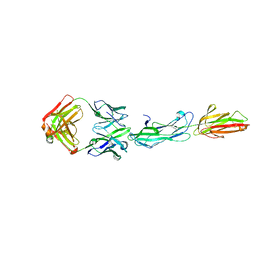 | | Identification of an Immunodominant region on a Group A Streptococcus T-antigen Reveals Temperature-Dependent Motion in Pili | | Descriptor: | CALCIUM ION, Mouse-Human Fab heavy chain, Mouse-Human Fab light chain, ... | | Authors: | Raynes, J.M, Young, P.G, Moreland, N.J. | | Deposit date: | 2022-11-14 | | Release date: | 2023-03-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Identification of an immunodominant region on a group A Streptococcus T-antigen reveals temperature-dependent motion in pili.
Virulence, 14, 2023
|
|
8F5L
 
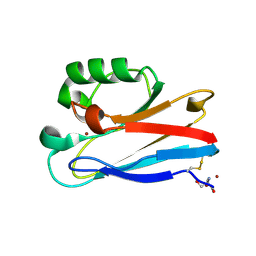 | | Azurin from Pseudomonas aeruginosa, Y72F/Y108F/F110L mutant | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Azurin, COPPER (II) ION | | Authors: | Zeug, M, Offenbacher, A.R, Choe, J. | | Deposit date: | 2022-11-14 | | Release date: | 2023-01-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Electrochemical and Structural Study of the Buried Tryptophan in Azurin: Effects of Hydration and Polarity on the Redox Potential of W48.
J.Phys.Chem.B, 127, 2023
|
|
8F5I
 
 | | SARS-CoV-2 S2 helix epitope scaffold bound by antibody DH1057.1 | | Descriptor: | DH1057.1 HC, DH1057.1 LC, Specialized acyl carrier protein, ... | | Authors: | Kapingidza, A.B, Wrapp, D, Winters, K, Azoitei, M.L. | | Deposit date: | 2022-11-14 | | Release date: | 2023-10-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Engineered immunogens to elicit antibodies against conserved coronavirus epitopes.
Nat Commun, 14, 2023
|
|
8F5H
 
 | |
8F5A
 
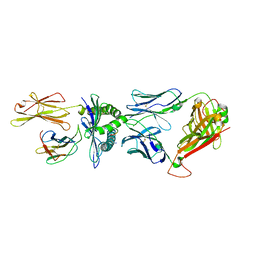 | |
8F59
 
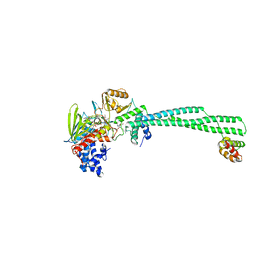 | | LSD1-CoREST in complex with AW2 and SNAG peptide | | Descriptor: | Lysine-specific histone demethylase 1A, REST corepressor 1, Zinc finger protein SNAI1, ... | | Authors: | Caroli, J, Mattevi, A. | | Deposit date: | 2022-11-12 | | Release date: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Distal drug resistance mutations promote covalent inhibitor-adduct Grob fragmentation in LSD1
to be published
|
|
8F58
 
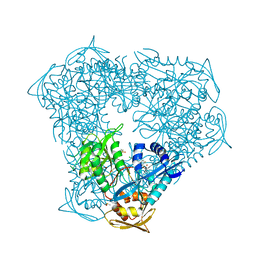 | | Crystal structure of acetyltransferase Eis from M. tuberculosis in complex with inhibitor SGT1616 | | Descriptor: | (1E)-1-[(4-chlorophenyl)methylidene]-2-(4-fluorophenyl)hydrazine, DI(HYDROXYETHYL)ETHER, DIMETHYL SULFOXIDE, ... | | Authors: | Pang, A.H, Punetha, A, Garneau-Tsodikova, S, Tsodikov, O.V. | | Deposit date: | 2022-11-12 | | Release date: | 2023-02-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery and Mechanistic Analysis of Structurally Diverse Inhibitors of Acetyltransferase Eis among FDA-Approved Drugs.
Biochemistry, 62, 2023
|
|
8F57
 
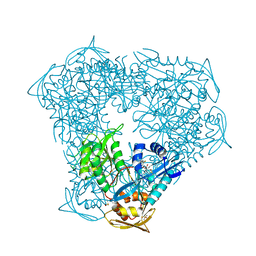 | | Crystal structure of acetyltransferase Eis from M. tuberculosis in complex with inhibitor SGT1615 | | Descriptor: | (1E)-1-[(4-chlorophenyl)methylidene]-2-(3-fluorophenyl)hydrazine, DI(HYDROXYETHYL)ETHER, DIMETHYL SULFOXIDE, ... | | Authors: | Pang, A.H, Punetha, A, Garneau-Tsodikova, S, Tsodikov, O.V. | | Deposit date: | 2022-11-12 | | Release date: | 2023-02-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Discovery and Mechanistic Analysis of Structurally Diverse Inhibitors of Acetyltransferase Eis among FDA-Approved Drugs.
Biochemistry, 62, 2023
|
|
8F55
 
 | | Crystal structure of acetyltransferase Eis from Mycobacterium tuberculosis H37Rv in complex with inhibitor SGT1614 | | Descriptor: | (2E)-2-[(4-chlorophenyl)methylidene]hydrazine-1-carboximidamide, DI(HYDROXYETHYL)ETHER, DIMETHYL SULFOXIDE, ... | | Authors: | Pang, A.H, Punetha, A, Garneau-Tsodikova, S, Tsodikov, O.V. | | Deposit date: | 2022-11-11 | | Release date: | 2023-02-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery and Mechanistic Analysis of Structurally Diverse Inhibitors of Acetyltransferase Eis among FDA-Approved Drugs.
Biochemistry, 62, 2023
|
|
8F51
 
 | | Crystal structure of acetyltransferase Eis from M. tuberculosis in complex with mefloquine | | Descriptor: | DI(HYDROXYETHYL)ETHER, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Pang, A.H, Punetha, A, Garneau-Tsodikova, S, Tsodikov, O.V. | | Deposit date: | 2022-11-11 | | Release date: | 2023-02-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Discovery and Mechanistic Analysis of Structurally Diverse Inhibitors of Acetyltransferase Eis among FDA-Approved Drugs.
Biochemistry, 62, 2023
|
|
8F4Z
 
 | | Crystal structure of acetyltransferase Eis from M. tuberculosis in complex with chloroquine | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, N-acetyltransferase Eis, ... | | Authors: | Pang, A.H, Punetha, A, Garneau-Tsodikova, S, Tsodikov, O.V. | | Deposit date: | 2022-11-11 | | Release date: | 2023-02-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery and Mechanistic Analysis of Structurally Diverse Inhibitors of Acetyltransferase Eis among FDA-Approved Drugs.
Biochemistry, 62, 2023
|
|
8F4Y
 
 | | Crystal Structure of SARS-CoV-2 2'-O-Methyltransferase in Complex with Compound 5a covalently bound to nsp16 and nsp10 | | Descriptor: | 2'-O-methyltransferase, 4-[(E)-2-(2,4-dichlorophenyl)ethenyl]-6-(trifluoromethyl)pyrimidin-2-ol, 4-[2-(2,4-dichlorophenyl)ethyl]-6-(trifluoromethyl)pyrimidin-2-ol, ... | | Authors: | Minasov, G, Shuvalova, L, Brunzelle, J.S, Rosas-Lemus, M, Kiryukhina, O, Satchell, K.J.F, Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2022-11-11 | | Release date: | 2023-10-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Discovery of a Druggable, Cryptic Pocket in SARS-CoV-2 nsp16 Using Allosteric Inhibitors.
Acs Infect Dis., 9, 2023
|
|
8F4W
 
 | | Crystal structure of acetyltransferase Eis from M. tuberculosis in complex with venlafaxine | | Descriptor: | 1-[(1R)-2-(dimethylamino)-1-(4-methoxyphenyl)ethyl]cyclohexan-1-ol, DI(HYDROXYETHYL)ETHER, DIMETHYL SULFOXIDE, ... | | Authors: | Pang, A.H, Punetha, A, Garneau-Tsodikova, S, Tsodikov, O.V. | | Deposit date: | 2022-11-11 | | Release date: | 2023-02-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery and Mechanistic Analysis of Structurally Diverse Inhibitors of Acetyltransferase Eis among FDA-Approved Drugs.
Biochemistry, 62, 2023
|
|
8F4V
 
 | | Alpha7 nicotinic acetylcholine receptor intracellular and transmembrane domains bound to ivermectin in a desensitized state | | Descriptor: | (2aE,4E,5'S,6S,6'R,7S,8E,11R,13R,15S,17aR,20R,20aR,20bS)-6'-[(2S)-butan-2-yl]-20,20b-dihydroxy-5',6,8,19-tetramethyl-17 -oxo-3',4',5',6,6',10,11,14,15,17,17a,20,20a,20b-tetradecahydro-2H,7H-spiro[11,15-methanofuro[4,3,2-pq][2,6]benzodioxacy clooctadecine-13,2'-pyran]-7-yl 2,6-dideoxy-4-O-(2,6-dideoxy-3-O-methyl-alpha-L-arabino-hexopyranosyl)-3-O-methyl-alpha-L-arabino-hexopyranoside, Neuronal acetylcholine receptor subunit alpha-7 | | Authors: | Bondarenko, V, Chen, Q, Tang, P. | | Deposit date: | 2022-11-11 | | Release date: | 2023-02-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Elucidation of Ivermectin Binding to alpha 7nAChR and the Induced Channel Desensitization.
Acs Chem Neurosci, 14, 2023
|
|
