1KCF
 
 | | Crystal Structure of the Yeast Mitochondrial Holliday Junction Resolvase, Ydc2 | | Descriptor: | HYPOTHETICAL 30.2 KD PROTEIN C25G10.02 IN CHROMOSOME I, SULFATE ION | | Authors: | Ceschini, S, Keeley, A, McAlister, M.S.B, Oram, M, Phelan, J, Pearl, L.H, Tsaneva, I.R, Barrett, T.E. | | Deposit date: | 2001-11-08 | | Release date: | 2001-11-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the fission yeast mitochondrial Holliday junction resolvase Ydc2.
EMBO J., 20, 2001
|
|
1KCI
 
 | | Crystal Structure of 9-amino-N-[2-(4-morpholinyl)ethyl]-4-acridinecarboxamide Bound to d(CGTACG)2 | | Descriptor: | 5'-D(*CP*GP*TP*AP*CP*G)-3', 9-AMINO-N-[2-(4-MORPHOLINYL)ETHYL]-4-ACRIDINECARBOXAMIDE | | Authors: | Adams, A, Guss, J.M, Denny, W.A, Wakelin, L.P.G. | | Deposit date: | 2001-11-08 | | Release date: | 2002-02-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of 9-amino-N-[2-(4-morpholinyl)ethyl]-4-acridinecarboxamide bound to d(CGTACG)2: implications for structure-activity relationships of acridinecarboxamide topoisomerase poisons.
Nucleic Acids Res., 30, 2002
|
|
7A7H
 
 | | Crystal structure of PPARgamma in complex with compound TK90 | | Descriptor: | (2~{R})-2-[[4-[[4-methoxy-2-(trifluoromethyl)phenyl]methylcarbamoyl]phenyl]methyl]butanoic acid, 1,2-ETHANEDIOL, Peroxisome proliferator-activated receptor gamma | | Authors: | Ni, X, Kirchner, T, Proschak, E, Chaikuad, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-08-28 | | Release date: | 2021-08-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Combined Cardioprotective and Adipocyte Browning Effects Promoted by the Eutomer of Dual sEH/PPAR gamma Modulator.
J.Med.Chem., 64, 2021
|
|
6ZW2
 
 | | Crystal structure of OXA-10loop48 in complex with hydrolyzed meropenem | | Descriptor: | (2S,3R)-2-[(2S,3R)-1,3-bis(oxidanyl)-1-oxidanylidene-butan-2-yl]-4-[(3S,5S)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfan yl-3-methyl-2,3-dihydro-1H-pyrrole-5-carboxylic acid, 1,2-ETHANEDIOL, Beta-lactamase, ... | | Authors: | Tassone, G, Di Pisa, F, Benvenuti, M, De Luca, F, Pozzi, C, Mangani, S, Docquier, J.D. | | Deposit date: | 2020-07-27 | | Release date: | 2021-08-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Mechanistic insights into carbapenem hydrolysis by OXA-48 and the OXA10-derived hybrids OXA-10 loop24 and loop48
To Be Published
|
|
1KCG
 
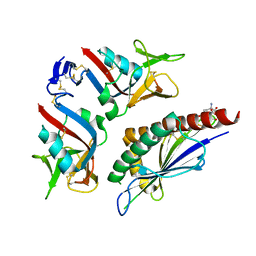 | | NKG2D in complex with ULBP3 | | Descriptor: | NKG2-D type II integral membrane protein, UL16-binding protein 3 | | Authors: | Radaev, S, Sun, P. | | Deposit date: | 2001-11-08 | | Release date: | 2002-01-09 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Conformational plasticity revealed by the cocrystal structure of NKG2D and its class I MHC-like ligand ULBP3.
Immunity, 15, 2001
|
|
1KLN
 
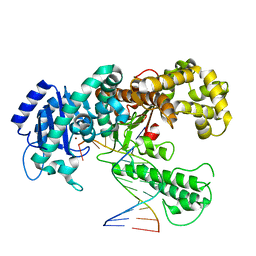 | | DNA POLYMERASE I KLENOW FRAGMENT (E.C.2.7.7.7) MUTANT/DNA COMPLEX | | Descriptor: | DNA (5'-D(*GP*CP*CP*GP*CP*GP*AP*GP*GP*C)-3'), DNA (5'-D(*GP*CP*CP*TP*CP*GP*CP*GP*GP*CP*GP*GP*C)-3'), PROTEIN (DNA POLYMERASE I KLENOW FRAGMENT (E.C.2.7.7.7)), ... | | Authors: | Beese, L.S, Derbyshire, V, Steitz, T.A. | | Deposit date: | 1994-05-24 | | Release date: | 1994-11-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure of DNA polymerase I Klenow fragment bound to duplex DNA.
Science, 260, 1993
|
|
1KM3
 
 | |
1KMB
 
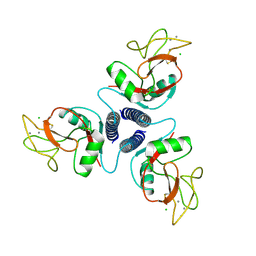 | | SELECTIN-LIKE MUTANT OF MANNOSE-BINDING PROTEIN A | | Descriptor: | CALCIUM ION, CHLORIDE ION, MANNOSE-BINDING PROTEIN-A | | Authors: | Ng, K.K.-S, Weis, W.I. | | Deposit date: | 1996-11-07 | | Release date: | 1997-02-12 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of a selectin-like mutant of mannose-binding protein complexed with sialylated and sulfated Lewis(x) oligosaccharides.
Biochemistry, 36, 1997
|
|
1KAJ
 
 | |
1KCM
 
 | | Crystal Structure of Mouse PITP Alpha Void of Bound Phospholipid at 2.0 Angstroms Resolution | | Descriptor: | Phosphatidylinositol Transfer Protein alpha | | Authors: | Schouten, A, Agianian, B, Westerman, J, Kroon, J, Wirtz, K.W.A, Gros, P. | | Deposit date: | 2001-11-09 | | Release date: | 2002-05-08 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of apo-phosphatidylinositol transfer protein alpha provides insight into membrane association.
EMBO J., 21, 2002
|
|
1KCQ
 
 | | Human Gelsolin Domain 2 with a Cd2+ bound | | Descriptor: | CADMIUM ION, GELSOLIN | | Authors: | Kazmirski, S.L, Isaacson, R.L, An, C, Buckle, A, Johnson, C.M, Daggett, V, Fersht, A.R. | | Deposit date: | 2001-11-09 | | Release date: | 2002-01-04 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Loss of a metal-binding site in gelsolin leads to familial amyloidosis-Finnish type.
Nat.Struct.Biol., 9, 2002
|
|
1KNF
 
 | | Crystal Structure of 2,3-dihydroxybiphenyl 1,2-dioxygenase Complexed with 3-methyl Catechol under Anaerobic Condition | | Descriptor: | 2,3-DIHYDROXYBIPHENYL 1,2-DIOXYGENASE, 3-METHYLCATECHOL, FE (II) ION, ... | | Authors: | Han, S, Bolin, J.T. | | Deposit date: | 2001-12-18 | | Release date: | 2002-03-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular basis for the stabilization and inhibition of 2, 3-dihydroxybiphenyl 1,2-dioxygenase by t-butanol.
J.Biol.Chem., 273, 1998
|
|
1KO1
 
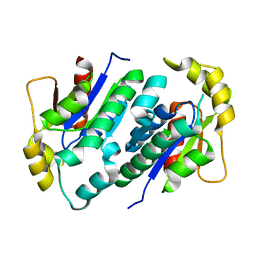 | | Crystal structure of gluconate kinase | | Descriptor: | CHLORIDE ION, Gluconate kinase | | Authors: | Kraft, L, Sprenger, G.A, Lindqvist, Y. | | Deposit date: | 2001-12-20 | | Release date: | 2002-05-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Conformational changes during the catalytic cycle of gluconate kinase as revealed by X-ray crystallography.
J.Mol.Biol., 318, 2002
|
|
1KOY
 
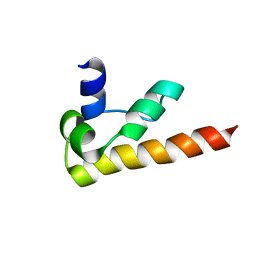 | | NMR structure of DFF-C domain | | Descriptor: | DNA fragmentation factor alpha subunit | | Authors: | Fukushima, K, Kikuchi, J, Koshiba, S, Kigawa, T, Kuroda, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2001-12-25 | | Release date: | 2002-09-04 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the DFF-C domain of DFF45/ICAD. A structural basis for the regulation of apoptotic DNA fragmentation.
J.Mol.Biol., 321, 2002
|
|
1K8A
 
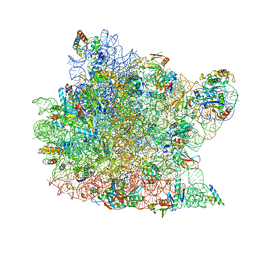 | | Co-crystal structure of Carbomycin A bound to the 50S ribosomal subunit of Haloarcula marismortui | | Descriptor: | 23S RRNA, 5S RRNA, CADMIUM ION, ... | | Authors: | Hansen, J.L, Ippolito, J.A, Ban, N, Nissen, P, Moore, P.B, Steitz, T. | | Deposit date: | 2001-10-23 | | Release date: | 2002-07-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The structures of four macrolide antibiotics bound to the large ribosomal subunit.
Mol.Cell, 10, 2002
|
|
6ZYU
 
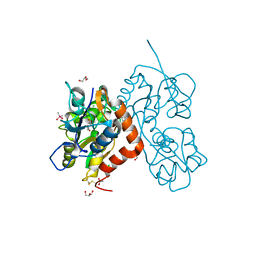 | | Structure of the GluA2 ligand-binding domain (L483Y-N754S) in complex with glutamate and BPAM549 | | Descriptor: | (4~{R})-4-cyclopropyl-7-fluoranyl-3,4-dihydro-2~{H}-thiochromene 1,1-dioxide, (4~{S})-4-cyclopropyl-7-fluoranyl-3,4-dihydro-2~{H}-thiochromene 1,1-dioxide, ACETATE ION, ... | | Authors: | Dorosz, J, Christensen, K.M, Kastrup, J.S. | | Deposit date: | 2020-08-03 | | Release date: | 2021-08-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Development of Thiochroman Dioxide Analogues of Benzothiadiazine Dioxides as New Positive Allosteric Modulators of alpha-Amino-3-hydroxy-5-methyl-4-isoxazolepropionic Acid (AMPA) Receptors.
Acs Chem Neurosci, 12, 2021
|
|
1KP3
 
 | |
1KPH
 
 | | Crystal Structure of mycolic acid cyclopropane synthase CmaA1 complexed with SAH and DDDMAB | | Descriptor: | CARBONATE ION, CYCLOPROPANE-FATTY-ACYL-PHOSPHOLIPID SYNTHASE 1, DIDECYL-DIMETHYL-AMMONIUM, ... | | Authors: | Huang, C.-C, Smith, C.V, Jacobs Jr, W.R, Glickman, M.S, Sacchettini, J.C, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2001-12-30 | | Release date: | 2002-01-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of mycolic acid cyclopropane synthases from Mycobacterium tuberculosis
J.Biol.Chem., 277, 2002
|
|
1KQA
 
 | |
1KQN
 
 | | Crystal structure of NMN/NaMN adenylyltransferase complexed with NAD | | Descriptor: | NICOTINAMIDE MONONUCLEOTIDE ADENYLYL TRANSFERASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, XENON | | Authors: | Zhou, T, Kurnasov, O, Tomchick, D.R, Binns, D.D, Grishin, N.V, Marquez, V.E, Osterman, A.L, Zhang, H. | | Deposit date: | 2002-01-07 | | Release date: | 2003-01-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of Human Nicotinamide/Nicotonic Acid Mononucleotide Adenylyltransferase. Basis for the dual substrate specificity and activation of the oncolytic agent tiazofurin.
J.Biol.Chem., 277, 2003
|
|
1KRQ
 
 | | CRYSTAL STRUCTURE ANALYSIS OF CAMPYLOBACTER JEJUNI FERRITIN | | Descriptor: | ferritin | | Authors: | Hortolan, L, Saintout, N, Granier, G, Langlois d'Estaintot, B, Manigand, C, Mizunoe, Y, Wai, S.N, Gallois, B, Precigoux, G. | | Deposit date: | 2002-01-10 | | Release date: | 2002-02-06 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | STRUCTURE OF CAMPYLOBACTER JEJUNI FERRITIN AT 2.7 A RESOLUTION
To be Published
|
|
1KRX
 
 | | SOLUTION STRUCTURE OF BERYLLOFLUORIDE-ACTIVATED NTRC RECEIVER DOMAIN: MODEL STRUCTURES INCORPORATING ACTIVE SITE CONTACTS | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, NITROGEN REGULATION PROTEIN NR(I) | | Authors: | Hastings, C.A, Lee, S.-Y, Cho, H.S, Yan, D, Kustu, S, Wemmer, D.E. | | Deposit date: | 2002-01-10 | | Release date: | 2003-08-19 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | High-Resolution Solution Structure of the Beryllofluoride-Activated NtrC Receiver Domain
Biochemistry, 42, 2003
|
|
7A36
 
 | |
7YZM
 
 | | MgADPNP-bound DCCP:DCCP-R complex | | Descriptor: | (R,R)-2,3-BUTANEDIOL, AMMONIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Jeoung, J.-H, Dobbek, H. | | Deposit date: | 2022-02-21 | | Release date: | 2022-12-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structural basis for coupled ATP-driven electron transfer in the double-cubane cluster protein.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7A3W
 
 | | Structure of Imine Reductase from Pseudomonas sp. | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, NAD(P)-dependent oxidoreductase, ... | | Authors: | Cuetos, A, Thorpe, T, Turner, N.J, Grogan, G. | | Deposit date: | 2020-08-18 | | Release date: | 2021-08-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Multifunctional biocatalyst for conjugate reduction and reductive amination.
Nature, 604, 2022
|
|
