3VID
 
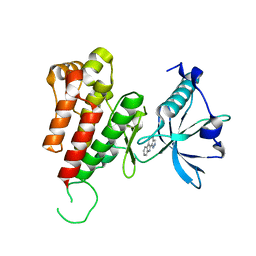 | | Crystal structure of human VEGFR2 kinase domain with Compound A. | | Descriptor: | 4,5,6,11-tetrahydro-1H-pyrazolo[4',3':6,7]cyclohepta[1,2-b]indole, Vascular endothelial growth factor receptor 2 | | Authors: | Iwata, H, Oki, H, Okada, K, Takagi, T, Tawada, M, Miyazaki, Y, Imamura, S, Hori, A, Hixon, M.S, Kimura, H, Miki, H. | | Deposit date: | 2011-09-29 | | Release date: | 2012-08-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A Back-to-Front Fragment-Based Drug Design Search Strategy Targeting the DFG-Out Pocket of Protein Tyrosine Kinases.
ACS MED.CHEM.LETT., 3, 2012
|
|
8YIH
 
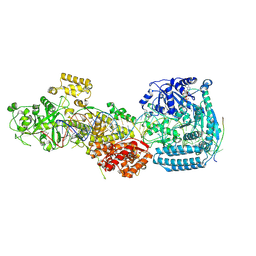 | | DmDcr-2/LoqsPD/slm1 in pre-dicing state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Dicer-2, isoform A, ... | | Authors: | Cao, N, Su, S, Wang, J, Ma, J, Wang, H.-W. | | Deposit date: | 2024-02-29 | | Release date: | 2025-03-05 | | Last modified: | 2025-03-12 | | Method: | ELECTRON MICROSCOPY (4.08 Å) | | Cite: | Structural basis of endo-siRNA processing by Drosophila Dicer-2 and Loqs-PD.
Nucleic Acids Res., 53, 2025
|
|
8YII
 
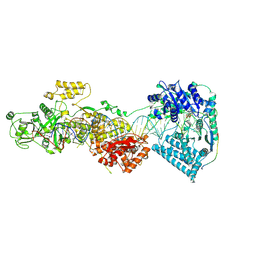 | | DmDcr-2/LoqsPD/slm2 in dicing state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Dicer-2, isoform A, ... | | Authors: | Cao, N, Su, S, Wang, J, Ma, J, Wang, H.-W. | | Deposit date: | 2024-02-29 | | Release date: | 2025-03-05 | | Last modified: | 2025-03-12 | | Method: | ELECTRON MICROSCOPY (3.42 Å) | | Cite: | Structural basis of endo-siRNA processing by Drosophila Dicer-2 and Loqs-PD.
Nucleic Acids Res., 53, 2025
|
|
5HNU
 
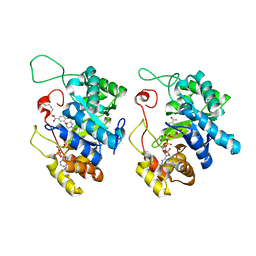 | | Crystal Structure of AKR1C3 complexed with octyl gallate | | Descriptor: | Aldo-keto reductase family 1 member C3, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, octyl 3,4,5-trihydroxybenzoate | | Authors: | Li, C, Zhao, Y, Zhang, H, Hu, X. | | Deposit date: | 2016-01-18 | | Release date: | 2017-01-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of AKR1C3 complexed with octyl gallte
To Be Published
|
|
6Y8K
 
 | | Crystal structure of CD137 in complex with the cyclic peptide BCY10916 | | Descriptor: | 1,1',1''-(1,3,5-triazinane-1,3,5-triyl)tripropan-1-one, 1,2-ETHANEDIOL, ACETATE ION, ... | | Authors: | Upadhyaya, P, Kublin, J, Dods, R, Kristensson, J, Lahdenranta, J, Kleyman, M, Repash, E, Ma, J, Mudd, G, Van Rietschoten, K, Haines, E, Harrison, H, Beswick, P, Chen, L, McDonnell, K, Battula, S, Hurov, K, Keen, N. | | Deposit date: | 2020-03-05 | | Release date: | 2021-02-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.011 Å) | | Cite: | Anticancer immunity induced by a synthetic tumor-targeted CD137 agonist.
J Immunother Cancer, 9, 2021
|
|
4EQH
 
 | |
1KJI
 
 | | Crystal structure of glycinamide ribonucleotide transformylase in complex with Mg-AMPPCP | | Descriptor: | 1,2-ETHANEDIOL, 3[N-MORPHOLINO]PROPANE SULFONIC ACID, CHLORIDE ION, ... | | Authors: | Thoden, J.B, Firestine, S.M, Benkovic, S.J, Holden, H.M. | | Deposit date: | 2001-12-04 | | Release date: | 2002-06-28 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | PurT-encoded glycinamide ribonucleotide transformylase. Accommodation of adenosine nucleotide analogs within the active site.
J.Biol.Chem., 277, 2002
|
|
4E5O
 
 | | Crystal structure of mouse thymidylate synthase in complex with dUMP | | Descriptor: | 1,4-BUTANEDIOL, 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, Thymidylate synthase | | Authors: | Dowiercial, A, Jarmula, A, Rypniewski, W, Sokolowska, M, Fraczyk, T, Ciesla, J, Rode, W. | | Deposit date: | 2012-03-14 | | Release date: | 2012-04-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mouse thymidylate synthase does not show the inactive conformation, observed for the human enzyme
Struct Chem, 2016
|
|
4NJU
 
 | | Crystal structure of multidrug-resistant clinical isolate A02 HIV-1 protease in complex with tipranavir | | Descriptor: | N-(3-{(1R)-1-[(6R)-4-HYDROXY-2-OXO-6-PHENETHYL-6-PROPYL-5,6-DIHYDRO-2H-PYRAN-3-YL]PROPYL}PHENYL)-5-(TRIFLUOROMETHYL)-2-PYRIDINESULFONAMIDE, Protease | | Authors: | Yedidi, R.S, Garimella, H, Kaufman, J.D, Das, D, Wingfield, P.T, Mitsuya, H. | | Deposit date: | 2013-11-11 | | Release date: | 2014-04-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A Conserved Hydrogen-Bonding Network of P2 bis-Tetrahydrofuran-Containing HIV-1 Protease Inhibitors (PIs) with a Protease Active-Site Amino Acid Backbone Aids in Their Activity against PI-Resistant HIV.
Antimicrob.Agents Chemother., 58, 2014
|
|
7XXH
 
 | | Cryo-EM structure of the purinergic receptor P2Y1R in complex with 2MeSADP and G11 | | Descriptor: | 2-(methylsulfanyl)adenosine 5'-(trihydrogen diphosphate), Guanine nucleotide-binding protein G(11) subunit alpha, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Tan, Q, Li, B, Han, S, Zhao, Q, Wu, B. | | Deposit date: | 2022-05-30 | | Release date: | 2023-06-07 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural insights into signal transduction of the purinergic receptors P2Y1R and P2Y12R.
Protein Cell, 14, 2023
|
|
5VYU
 
 | | Crystal structure of the WbkC N-formyltransferase from Brucella melitensis in complex with GDP-perosaminea and N-10-formyltetrahydrofolate | | Descriptor: | GDP-perosamine, GUANOSINE-5'-DIPHOSPHATE, Gdp-mannose 4,6-dehydratase / gdp-4-amino-4,6-dideoxy-d-mannose formyltransferase, ... | | Authors: | Riegert, A.S, Chantigian, D.P, Thoden, J.B, Holden, H.M. | | Deposit date: | 2017-05-26 | | Release date: | 2017-07-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Biochemical Characterization of WbkC, an N-Formyltransferase from Brucella melitensis.
Biochemistry, 56, 2017
|
|
3VQS
 
 | | Crystal structure of HCV NS5B RNA polymerase with a novel piperazine inhibitor | | Descriptor: | (2R)-4-(5-cyclopropyl[1,3]thiazolo[4,5-d]pyrimidin-2-yl)-N-[3-fluoro-4-(trifluoromethoxy)benzyl]-1-{[4-(trifluoromethyl)phenyl]sulfonyl}piperazine-2-carboxamide, CHLORIDE ION, RNA-directed RNA polymerase | | Authors: | Adachi, T, Doi, S, Ando, I, Sugimoto, K, Orita, T, Nomura, A, Kamada, M. | | Deposit date: | 2012-03-30 | | Release date: | 2012-08-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Preclinical Characterization of JTK-853, a Novel Nonnucleoside Inhibitor of the Hepatitis C Virus RNA-Dependent RNA Polymerase.
Antimicrob.Agents Chemother., 56, 2012
|
|
5ZGZ
 
 | |
4NMC
 
 | |
4NR4
 
 | | Crystal structure of the bromodomain of human CREBBP in complex with an isoxazolyl-benzimidazole ligand | | Descriptor: | 1,2-ETHANEDIOL, 1-(4-chlorobenzyl)-5-(3,5-dimethyl-1,2-oxazol-4-yl)-1H-benzimidazole, CREB-binding protein, ... | | Authors: | Filippakopoulos, P, Picaud, S, Felletar, I, Hay, D, Fedorov, O, Martin, S, von Delft, F, Brennan, P, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-11-26 | | Release date: | 2013-12-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Crystal structure of the bromodomain of human CREBBP in complex with an isoxazolyl-benzimidazole ligand
TO BE PUBLISHED
|
|
7P5Z
 
 | | Structure of a DNA-loaded MCM double hexamer engaged with the Dbf4-dependent kinase | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Cell division control protein 7, ... | | Authors: | Greiwe, J.F, Miller, T.C.R, Martino, F, Costa, A. | | Deposit date: | 2021-07-15 | | Release date: | 2022-02-02 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural mechanism for the selective phosphorylation of DNA-loaded MCM double hexamers by the Dbf4-dependent kinase.
Nat.Struct.Mol.Biol., 29, 2022
|
|
6CI5
 
 | | Crystal structure of the formyltransferase PseJ from Anoxybacillus kamchatkensis in complex with UDP-4,6-dideoxy-4-formamido-L-AltNAc and tetrahydrofolate | | Descriptor: | (2R,3R,4S,5R,6S)-3-(acetylamino)-5-(formylamino)-4-hydroxy-6-methyltetrahydro-2H-pyran-2-yl [(2R,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl dihydrogen diphosphate (non-preferred name), N-[4-({[(6R)-2-amino-4-oxo-3,4,5,6,7,8-hexahydropteridin-6-yl]methyl}amino)benzoyl]-L-glutamic acid, SULFATE ION, ... | | Authors: | Reimer, J.M, Harb, I, Schmeing, T.M. | | Deposit date: | 2018-02-23 | | Release date: | 2018-10-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.00003052 Å) | | Cite: | Structural Insight into a Novel Formyltransferase and Evolution to a Nonribosomal Peptide Synthetase Tailoring Domain.
ACS Chem. Biol., 13, 2018
|
|
3Q8F
 
 | | Crystal structure of 2-Fluorohistine labeled Protective Antigen (pH 5.8) | | Descriptor: | CALCIUM ION, Protective antigen, TETRAETHYLENE GLYCOL | | Authors: | Lovell, S, Battaile, K.P, Rajapaksha, M, Janowiak, B.E, Andra, K.K, Bann, J.G. | | Deposit date: | 2011-01-06 | | Release date: | 2012-02-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | pH effects on binding between the anthrax protective antigen and the host cellular receptor CMG2.
Protein Sci., 21, 2012
|
|
3VR1
 
 | | Crystal structure analysis of the translation factor RF3 | | Descriptor: | GUANOSINE-5',3'-TETRAPHOSPHATE, Peptide chain release factor 3 | | Authors: | Kihira, K, Shomura, Y, Shibata, N, Kitamura, M, Higuchi, Y. | | Deposit date: | 2012-04-03 | | Release date: | 2012-09-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure analysis of the translation factor RF3 (release factor 3)
Febs Lett., 586, 2012
|
|
3QL8
 
 | | CDK2 in complex with inhibitor JWS-6-260 | | Descriptor: | 1,2-ETHANEDIOL, 3-(4-amino-1,3,5-triazin-2-yl)-4-hydroxybenzonitrile, Cyclin-dependent kinase 2 | | Authors: | Betzi, S, Alam, R, Han, H, Becker, A, Schonbrunn, E. | | Deposit date: | 2011-02-02 | | Release date: | 2012-08-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-guided optimization of novel CDK2 inhibitors discovered by high-throughput screening
To be Published
|
|
8GH6
 
 | |
2Y37
 
 | | The discovery of novel, potent and highly selective inhibitors of inducible nitric oxide synthase (iNOS) | | Descriptor: | 2-[(1R)-3-amino-1-phenyl-propoxy]-4-chloro-benzonitrile, 5,6,7,8-TETRAHYDROBIOPTERIN, GLYCEROL, ... | | Authors: | Cheshire, D.R, Andrews, G, Beaton, H.G, Birkinshaw, T.N, Boughton-Smith, N, Connolly, S, Cook, T.R, Cooper, A, Cooper, S.L, Cox, D, Dixon, J, Gensmantel, N, Hamley, P.J, Harrison, R, Hartopp, P, Kack, H, Luker, T, Mete, A, Millichip, I, Nicholls, D.J, Pimm, A.D, St-Gallay, S.A, Wallace, A.V. | | Deposit date: | 2010-12-19 | | Release date: | 2011-04-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The Discovery of Novel, Potent and Highly Selective Inhibitors of Inducible Nitric Oxide Synthase (Inos).
Bioorg.Med.Chem.Lett., 21, 2011
|
|
5ZO7
 
 | | Kinesin spindle protein Eg5 in complex with STLC-type inhibitor PVEI0138 | | Descriptor: | (2R)-2-azanyl-3-[[2-(4-methoxyphenyl)-2-tricyclo[9.4.0.0^{3,8}]pentadeca-1(11),3,5,7,12,14-hexaenyl]sulfanyl]propanoic acid, ADENOSINE-5'-DIPHOSPHATE, Kinesin-like protein KIF11, ... | | Authors: | Yokoyama, H, Sato, K. | | Deposit date: | 2018-04-12 | | Release date: | 2018-10-10 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and Thermodynamic Basis of the Enhanced Interaction between Kinesin Spindle Protein Eg5 and STLC-type Inhibitors.
Acs Omega, 3, 2018
|
|
3N4L
 
 | | BACE-1 in complex with ELN380842 | | Descriptor: | Beta-secretase 1, N-[(1S,2R)-1-(3,5-difluorobenzyl)-2-hydroxy-3-({1-[3-(1H-pyrazol-1-yl)phenyl]cyclohexyl}amino)propyl]acetamide | | Authors: | Yao, N.H. | | Deposit date: | 2010-05-21 | | Release date: | 2010-11-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Improving the permeability of the hydroxyethylamine BACE-1 inhibitors: structure-activity relationship of P2' substituents.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
5HRF
 
 | |
