4FK3
 
 | | B-Raf Kinase V600E Oncogenic Mutant in Complex with PLX3203 | | Descriptor: | N-{2,4-difluoro-3-[(5-pyridin-3-yl-1H-pyrrolo[2,3-b]pyridin-3-yl)carbonyl]phenyl}ethanesulfonamide, Serine/threonine-protein kinase B-raf | | Authors: | Zhang, Y, Wang, W, Zhang, K.Y.J. | | Deposit date: | 2012-06-12 | | Release date: | 2012-08-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Discovery of a selective inhibitor of oncogenic B-Raf kinase with potent antimelanoma activity.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
7O25
 
 | | Complex-B bound [FeFe]-hydrogenase maturase HydE from T. maritima (reaction triggered in the crystal) | | Descriptor: | 1,2-ETHANEDIOL, 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, CARBON MONOXIDE, ... | | Authors: | Rohac, R, Martin, L, Liu, L, Basu, D, Tao, L, Britt, R.D, Rauchfuss, T, Nicolet, Y. | | Deposit date: | 2021-03-30 | | Release date: | 2021-05-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Crystal Structure of the [FeFe]-Hydrogenase Maturase HydE Bound to Complex-B.
J.Am.Chem.Soc., 143, 2021
|
|
7O26
 
 | | Complex-B bound [FeFe]-hydrogenase maturase HydE fromT. Maritima (5'dA + Methionine) | | Descriptor: | 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, 5'-DEOXYADENOSINE, CARBON MONOXIDE, ... | | Authors: | Rohac, R, Martin, L, Liu, L, Basu, D, Tao, L, Britt, R.D, Rauchfuss, T, Nicolet, Y. | | Deposit date: | 2021-03-30 | | Release date: | 2021-05-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structure of the [FeFe]-Hydrogenase Maturase HydE Bound to Complex-B.
J.Am.Chem.Soc., 143, 2021
|
|
7O1S
 
 | | Complex-B bound [FeFe]-hydrogenase maturase HydE fromT. Maritima (Wild-type protein) | | Descriptor: | 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, CARBON MONOXIDE, CYANIDE ION, ... | | Authors: | Rohac, R, Martin, L, Liu, L, Basu, D, Tao, L, Britt, R.D, Rauchfuss, T, Nicolet, Y. | | Deposit date: | 2021-03-30 | | Release date: | 2021-05-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Crystal Structure of the [FeFe]-Hydrogenase Maturase HydE Bound to Complex-B.
J.Am.Chem.Soc., 143, 2021
|
|
7O1O
 
 | | Complex-B bound [FeFe]-hydrogenase maturase HydE fromT. Maritima (Auxiliary cluster deleted variant) | | Descriptor: | CARBON MONOXIDE, CHLORIDE ION, CYANIDE ION, ... | | Authors: | Rohac, R, Martin, L, Liu, L, Basu, D, Tao, L, Britt, R.D, Rauchfuss, T, Nicolet, Y. | | Deposit date: | 2021-03-30 | | Release date: | 2021-05-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Crystal Structure of the [FeFe]-Hydrogenase Maturase HydE Bound to Complex-B.
J.Am.Chem.Soc., 143, 2021
|
|
4UYN
 
 | | SAR156497 an exquisitely selective inhibitor of Aurora kinases | | Descriptor: | AURORA KINASE A, ethyl (9S)-9-[5-(1H-benzimidazol-2-ylsulfanyl)furan-2-yl]-8-hydroxy-5,6,7,9-tetrahydro-2H-pyrrolo[3,4-b]quinoline-3-carboxylate | | Authors: | Carry, J.C, Clerc, F, Minoux, H, Schio, L, Mauger, J, Nair, A, Parmantier, E, Lemoigne, R, Delorme, C, Nicolas, J.P, Krick, A, Abecassis, P.Y, Crocq-Stuerga, V, Pouzieux, S, Delarbre, L, Maignan, S, Bertrand, T, Bjergarde, K, Ma, N, Lachaud, S, Guizani, H, Lebel, R, Doerflinger, G, Monget, S, Perron, S, Gasse, F, Angouillant-Boniface, O, Filoche-Romme, B, Murer, M, Gontier, S, Prevost, C, Monteiro, M.L, Combeau, C. | | Deposit date: | 2014-09-02 | | Release date: | 2014-11-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Sar156497, an Exquisitely Selective Inhibitor of Aurora Kinases.
J.Med.Chem., 58, 2015
|
|
9J6K
 
 | | HBx complexed with DDB1 | | Descriptor: | DNA damage-binding protein 1, Protein X | | Authors: | Tanaka, H, Kita, S, Sasaki, M, Maenaka, K, Machida, S. | | Deposit date: | 2024-08-16 | | Release date: | 2025-06-04 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (2.68 Å) | | Cite: | Structural basis of the hepatitis B virus X protein in complex with DDB1.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
8RDF
 
 | |
8RDA
 
 | |
9L5T
 
 | | Cryo-EM structure of the thermophile spliceosome (state B*Q2) | | Descriptor: | Anaphase-promoting complex subunit 4-like WD40 domain-containing protein, CCDC12, Delta(14)-sterol reductase, ... | | Authors: | Li, Y, Fischer, P, Wang, M, Yuan, R, Meng, W, Luehrmann, R, Lau, B, Hurt, E, Cheng, J. | | Deposit date: | 2024-12-23 | | Release date: | 2025-03-12 | | Last modified: | 2025-04-16 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural insights into spliceosome fidelity: DHX35-GPATCH1- mediated rejection of aberrant splicing substrates.
Cell Res., 35, 2025
|
|
9L5S
 
 | | Cryo-EM structure of the thermophile spliceosome (state B*Q1) | | Descriptor: | Anaphase-promoting complex subunit 4-like WD40 domain-containing protein, CCDC12, Delta(14)-sterol reductase, ... | | Authors: | Li, Y, Fischer, P, Wang, M, Yuan, R, Meng, W, Luehrmann, R, Lau, B, Hurt, E, Cheng, J. | | Deposit date: | 2024-12-23 | | Release date: | 2025-03-12 | | Last modified: | 2025-04-16 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural insights into spliceosome fidelity: DHX35-GPATCH1- mediated rejection of aberrant splicing substrates.
Cell Res., 35, 2025
|
|
4LVG
 
 | | Fragment-based Identification of Amides Derived From trans-2-(Pyridin-3-yl)cyclopropanecarboxylic Acid as Potent Inhibitors of Human Nicotinamide Phosphoribosyltransferase (NAMPT) | | Descriptor: | (1S,2S)-N-[4-(phenylsulfonyl)phenyl]-2-(pyridin-3-yl)cyclopropanecarboxamide, 1,2-ETHANEDIOL, Nicotinamide phosphoribosyltransferase, ... | | Authors: | Giannetti, A.M, Zheng, X, Skelton, N, Wang, W, Bravo, B, Feng, Y, Gunzner-Toste, J, Ho, Y, Hua, R, Wang, C, Zhao, Q, Liederer, B.M, Liu, Y, O'Brien, T, Oeh, J, Sampath, D, Shen, Y, Wang, L, Wu, H, Xiao, Y, Yuen, P, Zak, M, Zhao, G, Dragovich, P.S. | | Deposit date: | 2013-07-26 | | Release date: | 2013-09-25 | | Last modified: | 2025-05-07 | | Method: | X-RAY DIFFRACTION (1.702 Å) | | Cite: | Identification of amides derived from 1H-pyrazolo[3,4-b]pyridine-5-carboxylic acid as potent inhibitors of human nicotinamide phosphoribosyltransferase (NAMPT).
Bioorg.Med.Chem.Lett., 23, 2013
|
|
9LYO
 
 | | Alpha SARS-CoV-2 spike protein in complex with REGN10987 Fab homologue. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, REGN10987 Fab homologue (Heavy chain), ... | | Authors: | Kocharovskaya, M.V, Pichkur, E.B, Shenkarev, Z.O, Lyukmanova, E.N. | | Deposit date: | 2025-02-20 | | Release date: | 2025-04-02 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Structure and dynamics of Alpha B.1.1.7 SARS-CoV-2 S-protein in complex with Fab of neutralizing antibody REGN10987.
Biochem.Biophys.Res.Commun., 755, 2025
|
|
9N2B
 
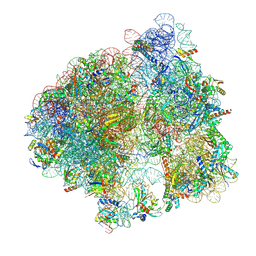 | | Impacts of ribosomal RNA sequence variation on gene expression and phenotype: Cryo-EM structure of the rrsB ribosome (BBB-70S) | | Descriptor: | 1,4-DIAMINOBUTANE, 16S ribosomal RNA (rRNA) from the rrnB operon, 23S ribosomal RNA (rRNA) from the rrnB operon, ... | | Authors: | Welfer, G.A, Brady, R.A, Natchiar, S.K, Watson, Z.L, Rundlet, E.J, Alejo, J.L, Singh, A.P, Mishra, N.K, Altman, R.B, Blanchard, S.C. | | Deposit date: | 2025-01-28 | | Release date: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | Impacts of ribosomal RNA sequence variation on gene expression and phenotype.
Philos.Trans.R.Soc.Lond.B Biol.Sci., 380, 2025
|
|
7R5X
 
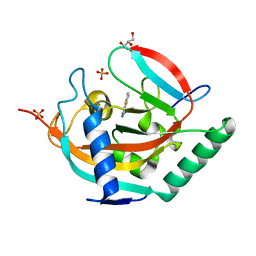 | | Tankyrase 2 in complex with an inhibitor (OUL211) | | Descriptor: | 7-methyl-[1,2,4]triazolo[3,4-b][1,3]benzothiazole, GLYCEROL, Poly [ADP-ribose] polymerase tankyrase-2, ... | | Authors: | Sowa, S.T, Lehtio, L. | | Deposit date: | 2022-02-11 | | Release date: | 2023-02-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.052 Å) | | Cite: | [1,2,4]Triazolo[3,4- b ]benzothiazole Scaffold as Versatile Nicotinamide Mimic Allowing Nanomolar Inhibition of Different PARP Enzymes.
J.Med.Chem., 66, 2023
|
|
2I7B
 
 | |
4V64
 
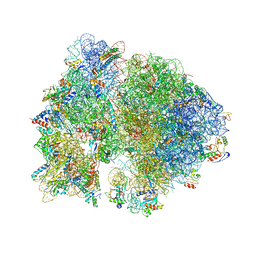 | | Crystal structure of the bacterial ribosome from Escherichia coli in complex with hygromycin B. | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Borovinskaya, M.A, Shoji, S, Fredrick, K, Cate, J.H.D. | | Deposit date: | 2008-06-11 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural basis for hygromycin B inhibition of protein biosynthesis
Rna, 14, 2008
|
|
1L2P
 
 | |
7R3O
 
 | | PARP15 catalytic domain in complex with OUL40 | | Descriptor: | 6-methyl-[1,2,4]triazolo[3,4-b][1,3]benzothiazole, DIMETHYL SULFOXIDE, Protein mono-ADP-ribosyltransferase PARP15 | | Authors: | Maksimainen, M.M, Murthy, S, Lehtio, L. | | Deposit date: | 2022-02-07 | | Release date: | 2023-01-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | [1,2,4]Triazolo[3,4- b ]benzothiazole Scaffold as Versatile Nicotinamide Mimic Allowing Nanomolar Inhibition of Different PARP Enzymes.
J.Med.Chem., 66, 2023
|
|
7R3L
 
 | | PARP14 catalytic domain in complex with OUL40 | | Descriptor: | 6-methyl-[1,2,4]triazolo[3,4-b][1,3]benzothiazole, CHLORIDE ION, Poly [ADP-ribose] polymerase 14, ... | | Authors: | Maksimainen, M.M, Murthy, S, Lehtio, L. | | Deposit date: | 2022-02-07 | | Release date: | 2023-01-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | [1,2,4]Triazolo[3,4- b ]benzothiazole Scaffold as Versatile Nicotinamide Mimic Allowing Nanomolar Inhibition of Different PARP Enzymes.
J.Med.Chem., 66, 2023
|
|
7R4A
 
 | | PARP15 catalytic domain in complex with OUL188 | | Descriptor: | 6,8-dimethyl-[1,2,4]triazolo[3,4-b][1,3]benzothiazole, DIMETHYL SULFOXIDE, Protein mono-ADP-ribosyltransferase PARP15 | | Authors: | Murthy, S, Maksimainen, M.M, Lehtio, L. | | Deposit date: | 2022-02-08 | | Release date: | 2023-01-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | [1,2,4]Triazolo[3,4- b ]benzothiazole Scaffold as Versatile Nicotinamide Mimic Allowing Nanomolar Inhibition of Different PARP Enzymes.
J.Med.Chem., 66, 2023
|
|
5DD3
 
 | |
5JPX
 
 | |
5DD5
 
 | |
8AAE
 
 | | CAII in complex with para-carboran-propylsulfonamid | | Descriptor: | Carbonic anhydrase 2, Para-Carborane propyl-sulfonamide, ZINC ION | | Authors: | Brynda, J, Rezacova, P. | | Deposit date: | 2022-07-01 | | Release date: | 2023-05-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | B-H⋯ pi and C-H⋯ pi interactions in protein-ligand complexes: carbonic anhydrase II inhibition by carborane sulfonamides.
Phys Chem Chem Phys, 25, 2023
|
|
